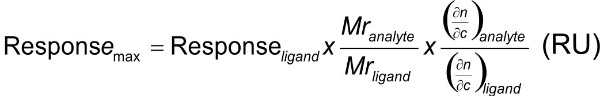This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit?
- NXTRay
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 3 weeks ago #1
by NXTRay
Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit? was created by NXTRay
Just asking, can we predict how low molecular weight (LMW) analyte our SPR system can detect based on our LOD (limit of detection) and our sensitivity on another protein detection? So right now our developed system can detect alpha-synuclein (14 kDa) with LOD around 5fg/ml. Can we predict how LMW can we detect? Is there any reference paper or something? Because as far as my knowledge we can get the change of RII based on the experiments first. But right now my case is I want to test the limit of our SPR devices to detect LMW (I'm choosing what LMW analytes to buy right now)
Thank you! any discussion or comments are very appreciated
Thank you! any discussion or comments are very appreciated
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 3 weeks ago #2
by Arnoud
Replied by Arnoud on topic Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit?
From the SPRpages book:
The binding of biomolecules results in the change of the refractive index on the sensor surface, which is measured as a change in SPR resonance angle or resonance wavelength. Fortunately, the change in refractive index on the surface is linear to the number of molecules bound to the sensor surface (1, 3). For the most used sensor surfaces (100 nm carboxymethyl dextran matrix) the relation between the amount of bound analyte and the resonance units is:
1000 RU = 10 mg/mL = 1 ng/mm2
And the maximum response can be predicted with:
However, this holds mainly for protein-protein interactions that have a refractive index increment (RII) of about 0.17–0.21 ml/g (1, 2, 4). For more specialized applications knowledge of the RII may be important for qualitative and comparative measurements, especially with small compounds (RII 0.15–0.34)(1). In the process of screening combinatorial libraries, many small molecules are screened at a single concentration. For an accurate affinity ranking and correct stoichiometric measurements, SPR response must be normalized for each compound (1). In addition, accurate RII values are required to estimate film thickness and surface coverage. An additional complication is the nature of the evanescent wave, which will provide different absolute values depending on the thickness of the sensor surface (2).
References:
1. Davis, T. M. and Wilson, W. D.; Determination of the Refractive Index Increments of Small Molecules for Correction of Surface Plasmon Resonance Data. Analytical Biochemistry (284) 2: 348-353; 2000.
2. Di Primo, C. and Lebars, I.; Determination of refractive index increment ratios for protein-nucleic acid complexes by surface plasmon resonance. Analytical Biochemistry (368) 2: 148-155; 2007.
3. Stenberg, E., Persson, B., Roos, H., et al.; Quantitative determination of surface concentration of protein with surface plasmon resonance by using radiolabelled proteins. Journal of Colloid and Interface Science (143) 2: 513-526; 1991.
4. Zhao, H., Brown, Patrick H. and Schuck, P.; On the Distribution of Protein Refractive Index Increments. Biophysical Journal (100) 9: 2309-2317; 2011.
I hope this will help you
kind regards
Arnoud
The binding of biomolecules results in the change of the refractive index on the sensor surface, which is measured as a change in SPR resonance angle or resonance wavelength. Fortunately, the change in refractive index on the surface is linear to the number of molecules bound to the sensor surface (1, 3). For the most used sensor surfaces (100 nm carboxymethyl dextran matrix) the relation between the amount of bound analyte and the resonance units is:
1000 RU = 10 mg/mL = 1 ng/mm2
And the maximum response can be predicted with:
However, this holds mainly for protein-protein interactions that have a refractive index increment (RII) of about 0.17–0.21 ml/g (1, 2, 4). For more specialized applications knowledge of the RII may be important for qualitative and comparative measurements, especially with small compounds (RII 0.15–0.34)(1). In the process of screening combinatorial libraries, many small molecules are screened at a single concentration. For an accurate affinity ranking and correct stoichiometric measurements, SPR response must be normalized for each compound (1). In addition, accurate RII values are required to estimate film thickness and surface coverage. An additional complication is the nature of the evanescent wave, which will provide different absolute values depending on the thickness of the sensor surface (2).
References:
1. Davis, T. M. and Wilson, W. D.; Determination of the Refractive Index Increments of Small Molecules for Correction of Surface Plasmon Resonance Data. Analytical Biochemistry (284) 2: 348-353; 2000.
2. Di Primo, C. and Lebars, I.; Determination of refractive index increment ratios for protein-nucleic acid complexes by surface plasmon resonance. Analytical Biochemistry (368) 2: 148-155; 2007.
3. Stenberg, E., Persson, B., Roos, H., et al.; Quantitative determination of surface concentration of protein with surface plasmon resonance by using radiolabelled proteins. Journal of Colloid and Interface Science (143) 2: 513-526; 1991.
4. Zhao, H., Brown, Patrick H. and Schuck, P.; On the Distribution of Protein Refractive Index Increments. Biophysical Journal (100) 9: 2309-2317; 2011.
I hope this will help you
kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- NXTRay
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 3 weeks ago #3
by NXTRay
Replied by NXTRay on topic Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit?
Thank you for your explanation,
But what if right now my condition is to choose which LMW protein my SPR can detect (I want to buy LMW protein), without knowing any responses for that LMW protein.
But what if right now my condition is to choose which LMW protein my SPR can detect (I want to buy LMW protein), without knowing any responses for that LMW protein.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 3 weeks ago #4
by Arnoud
Replied by Arnoud on topic Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit?
Currently you use an analyte of 15 kDa and you say that the LOD is 5 fg/ml. My question is: how much response do you get with your instrument? There must be a relation between analyte size, concentration and measured value. I never dealt with LOD's but when a signal is very low you can immobilize more ligand.
For instance when measuring interactions of small compounds to carbonic anhydrase you have to immobilize at least 3000 RU on a Biacore T200 to measure a compound of around 200 Da. So if you have calibrated (amount and size of ligand and concentration and size of analyte) your instrument you should be able to calculate the minimum MW of the analyte.
So the new set-up depends on the ligand and analyte sizes and the sensitivity of your SPR system.
1. Talibov, V. O., Linkuvienė, V., Matulis, D., et al.; Kinetically Selective Inhibitors of Human Carbonic Anhydrase Isozymes I, II, VII, IX, XII, and XIII. Journal of Medicinal Chemistry (59) 5: 2083-2093; 2016.
For instance when measuring interactions of small compounds to carbonic anhydrase you have to immobilize at least 3000 RU on a Biacore T200 to measure a compound of around 200 Da. So if you have calibrated (amount and size of ligand and concentration and size of analyte) your instrument you should be able to calculate the minimum MW of the analyte.
So the new set-up depends on the ligand and analyte sizes and the sensitivity of your SPR system.
1. Talibov, V. O., Linkuvienė, V., Matulis, D., et al.; Kinetically Selective Inhibitors of Human Carbonic Anhydrase Isozymes I, II, VII, IX, XII, and XIII. Journal of Medicinal Chemistry (59) 5: 2083-2093; 2016.
Please Log in or Create an account to join the conversation.
- NXTRay
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 3 weeks ago - 4 years 3 weeks ago #5
by NXTRay
Replied by NXTRay on topic Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit?
Thank you very much, I clearly understand now so its just a ratio between ligand and its analyte. But can it be used to define a ratio between known and unknown injected analyte? where A is my known analyte (alpha-syn spr response) and B is the new analyte I want to analyze
Last edit: 4 years 3 weeks ago by NXTRay.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 3 weeks ago #6
by Arnoud
Replied by Arnoud on topic Surface Plasmon Resonance (SPR) Predicting Molecular Weight Limit?
Yes, the mass ratio determines the response level.
That said you must have a binding of your new analyte to the ligand.
Arnoud
That said you must have a binding of your new analyte to the ligand.
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud