Low affinity / weak interactions / fast kinetics
Low affinity kinetics is characterized by fast on- and off-curves. Often the question is: Are they actual kinetics of the biomolecular interaction or is it bulk shift?
When it is actual low affinity kinetics of the biomolecular interaction, the shape of the curves is almost totally determined by the dissociation rate. Because of the fast dissociation, the curves reach equilibrium very quickly. The height of the response is directly proportional to the analyte concentration according to Langmuir equilibrium. With increasing analyte concentration, the surface will saturate as opposed to increasingly higher responses induced by high salt, DMSO, glycerol or non-specific binding and proper referencing is very important in this case.
Due to the fast association and dissociation kinetics, the rate constants of these interactions are difficult to calculate. Sometimes, a high sample rate can provide more data points to work with but in a lot of cases this will not be sufficient. In such situations, the only kinetic parameter that can be calculated is the equilibrium dissociation constant KD using equilibrium analysis software.
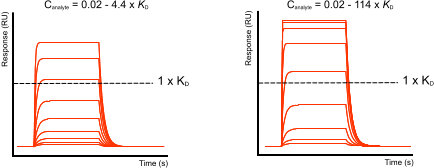
In the figure ‘Fast-on, fast-off curves’ there are two examples with the same fast kinetics but each with a different analyte concentration range. In the left example it is difficult to determine if the analyte will saturate the ligand. Therefore, high analyte concentrations are necessary to show that the ligand is saturating and that the maximal response level reaches a plateau.
Examples of fast kinetics can be found in the publications of Magotti et al. (1) where small peptides are tested for their inhibition quality on the complement system protein C3b and Souphron et al. (2) where the regulation of the ubiquitin selectivity is investigated.
In conclusion: real kinetics can saturate the ligand at high analyte concentrations. If the response keeps getting higher with increasing analyte concentrations, other effects such as non-specific binding or bulk refractive distortion can cause these high responses.
BiaSimulation
The BiaSimalution program is especially designed to explore slow and fast kinetics. The program can simulate all kinds of kinetics by varying the kinetic parameters such as ka, kd, analyte concentration and association and dissociation times. Please take a look at the SPR-simulation page.
References
| (1) | Magotti, P., D. Ricklin, H. Qu, et al. Structure-Kinetic Relationship Analysis of the Therapeutic Complement Inhibitor Compstatin. Journal of molecular recognition : JMR 22: 495-505; (2009). Goto reference |
| (2) | Souphron, J., M. B. Waddell, A. Paydar, et al. Structural Dissection of a Gating Mechanism Preventing Misactivation of Ubiquitin by NEDD8’s E1. Biochemistry 47: 8961-8969; (2008). Goto reference |