Purpose of the experiment
Depending on the application a known amount of ligand must be immobilized on the sensor chip surface. A crude guideline is the figure about immobilization levels.
- For specificity measurements, almost any ligand density will do as long as it gives a proper signal.
- Concentration measurements need the highest ligand density to facilitate mass transfer limitation. In a total mass transfer controlled experiment binding will depend on the analyte concentration and not on the binding kinetics between the ligand and analyte.
- Affinity ranking can be done with low to moderate density sensor chips. The important factor is that the analyte should saturate the ligand within a proper time frame.
- Kinetics are done with the lowest ligand density still giving a good response without being disturbed by secondary factors such as mass transfer or steric hindrance.
- Low molecular mass binding is done with high-density sensor chips to bind as much as possible of the component to gain a proper signal.
Some calculations
In general, for kinetic measurements, a total analyte response of maximal 100 RU, when the analyte is injected (1), (2) is desired. With this value in mind (Rmax), the amount of ligand (in response units) to be immobilized can be calculated with:

Or more conveniently:
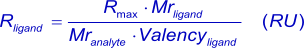
Because of the linear relation between response and amount of protein immobilized to the sensor surface (3), (4), the theoretical number of ligand sites after immobilization can be calculated (5).

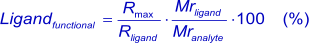
This formula is theoretical because it assumes that all immobilized ligand molecules are fully accessible and functional, which is most likely not the case with the standard amine coupling. When the Rmax is known after analyte injection, the percentage of functional ligand (the ligand that can bind the analyte) can be calculated. A low analyte response can be caused by low affinity, an impure ligand or that binding sites have been affected by the immobilization procedure. A to high response can be induced by non-specific binding or analyte aggregates (6).
Because the dextran matrix has a significant extension (in of the order of 100 nm for sensor chip CM5), "surface concentrations" on the sensor chip surface are strictly speaking volume concentrations. The relation between the actual protein 'concentration' and the Response units (RU) for the Biacore type machines (7), (8) is:

These relations make it possible to calculate the ligand concentration:

References
| (1) | Karlsson, R. et al Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. Journal of Immunological Methods 229-240; (1991). |
| (2) | Myszka, D. G. Survey of the 1998 optical biosensor literature. J.Mol.Recognit. 12: 390-408; (1999). |
| (3) | Stenberg, E. et al Quantitative determination of surface concentration of protein with surface plasmon resonance by using radiolabelled proteins. J.Colloid Interface Sci. 143: 513-526; (1991). |
| (4) | Quinn, J. G. et al Development and application of surface plasmon resonance-based biosensors for the detection of cell-ligand interactions. Analytical Biochemistry281: 135-143; (2000). |
| (5) | BIACORE AB BIACORE Technology Handbook. (1998). |
| (6) | BIACORE AB BIA Symposium '98. (1998). |
| (7) | BIACORE AB BIACORE Getting Started. (1998). |
| (8) | Muller, K. M. et al Model and simulation of multivalent binding to fixed ligands. Analytical Biochemistry 261: 149-158; (1998). |