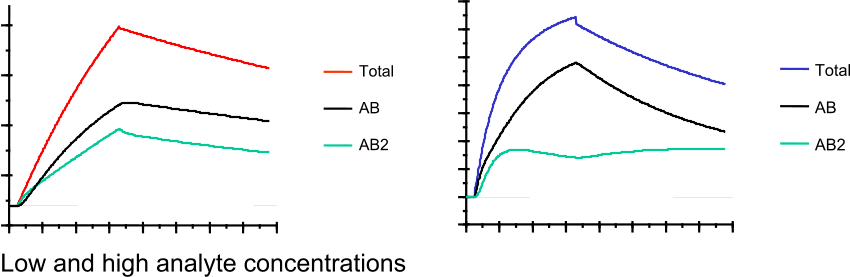Bivalent analyte
A number of analytes has two identical binding sites (e.g. an antibody). This allows the analyte to bind first with one site to the ligand. The other (free) site is now in closer contact with other ligand molecules and can bind to a second ligand site. The formation of the second binding will depend on the flexibility of the analyte, the type of ligand, the dextran layer and the availability of free ligand (1). In addition, the second binding will give a stabilization of the ligand-analyte complex without extra response and therefore shifts the equilibrium dissociation constant to a more stable interaction.
A bivalent analyte gives rise to two sets of rate constants, one for each binding step. The meaning of the two sets of rate constants and particular the second set of rate constants is difficult to interpret. The second association constant is dependent on the first binding (it is to some extent directed). In case of an antibody, both binding sites are equal, and consequently also the rate constants. However, since the second binding is dependent on the first interaction (it is to some extent directed) the second association rate constant is given in RU-1s-1.
In addition, the first and second kd are interchangeable. Depending on the values, the fit will be changed and the Rmax adjusted. Visual inspection of the sensorgram and matching the kd and Rmax with the curves seems the best option. To avoid this model, immobilize the bivalent analyte or use low ligand densities on for instance a plane sensor chip with no dextran layer.
Reaction equation
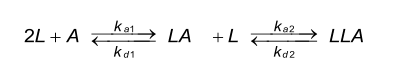
The factor of two is introduced because the first binding step can occur to either of the two unoccupied sites, while the second binding step can only occur to the remaining unoccupied site (2),(3). The binding model in Biacore differs from solution reactions because one molecule is immobilized.
Differential equation
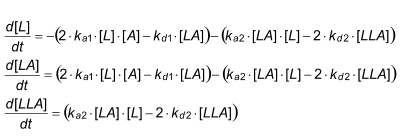
Parameters
L
concentration of free ligand in RU
A
concentration of free analyte in M
LA
concentration of ligand-analyte complex in RU
LAA
concentration of two ligand-analyte complex in RU
ka1
first association rate constant in M-1s-1
kd1
first dissociation rate constant in s-1
ka2
second association rate constant in RU-1s-1
kd2
second dissociation rate constant in s-1
Response calculation
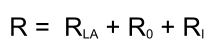
Below an example of a sensorgram generated with the differential equations from above.
| Concentration (nM) | ka1 (M-1s-1) | kd1 (s-1) | ka2 (RU-1s-1) | kd2 (s-1) | RMax (RU) |
|---|---|---|---|---|---|
| local | global | global | global | global | global |
| - | 1.105 | 5.10-3 | 1.10-4 | 0.1 | 250 |
| 12.5 | |||||
| 25 | |||||
| 50 | |||||
| 75 | |||||
| 100 | |||||
In case of a bivalent interaction, the dependency of the concentration on the components of the interaction can be studied. At low analyte concentrations, the LLA complex is dominating while at high analyte concentrations the LA complex is more abundant.
In general, the bivalent model works better for lower affinities when multivalency effects are easier to resolve. In order to resolve the kd of extremely stable interactions long dissociation times (hours, overnight) are needed (4). When comparing antibodies, it is better to capture the antibody under investigation with an anti Fc immobilized antibody. In this way, the bivalent nature of the antibody will not appear in the sensorgram.
In case of a CM5 sensor chip, the second association rate constant (ka2) in RU.s-1 can be converted to M-1s-1 with the following formula (5):
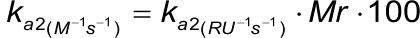
References
| (1) | Baumann, S., P. Grob, F. Stuart, et al. - Indirect immobilization of recombinant proteins to a solid phase using the albumin binding domain of streptococcal protein G and immobilized albumin. Journal of Immunological Methods 221: 95-106; (1998). |
| (2) | Nelson, R. W., J. W. Jarvik, B. E. Taillon, et al. - BIA/MS of epitope-tagged peptides directly from E. coli lysate: multiplex detection and protein identification at low-femtomole to subfemtomole levels. Analytical Chemistry 71: 2858-2865; (1999). |
| (3) | Cooper, M. A. and D. H. Williams - Kinetic analysis of antibody-antigen interactions at a supported lipid monolayer. Analytical Biochemistry 276: 36-47; (1999). |
| (4) | Biacore AB - Kinetic and Affinity analysis using BIA - Level 2. (1998). |
| (5) | Karlsson, R., J. A. Mo and R. Holmdahl - Binding of autoreactive mouse anti-type II collagen antibodies derived from the primary and the secondary immune response investigated with the biosensor technique. Journal of Immunological Methods 188: 63-71; (1995). |