Immobilization procedures
In general the immobilization procedure consists of three distinct parts.
- Activation: the priming of the sensor chip so it can form a covalent bond with another molecule
- Coupling: the injection of ligand so it froms covalent bonds with the sensor surface
- Deactivation: injection of a low molecular recative group to quence the remaining active surface groups
Each step has some points of attention.
Activation
In case of a covalent amine binding chemistry on the dextran-based sensor chips, the carboxyl groups are activated with a mixture of NHS (N-hydroxysuccinimide) and EDC (N-ethyl-N’-(dimethylaminopropyl) carbodiimide) to create N-Hydroxysuccinimide esters. By varying the activation time, more or fewer carboxyl groups are activated. In addition, the concentration of the NHS/EDC mixture can be varied to control the quantity of activated carboxyl groups. The quantity of activated groups determines how much ligand can bind to the sensor surface. The standard activation period for a CM5 sensor chip of BIACORE is 7 minutes with 0.05 M NHS/0.2M EDC at a flow rate of 5 µl/min.
Coupling
The reactivity of the ligand at the chosen pH determines how fast the ligand will bind to the activated surface. The rate of pre-concentration is directly related to the ligand concentration and pH of the immobilization solution. Ligand concentrations that are too high will give high ligand pre-concentration response but will also make it difficult to immobilize the proper amount of ligand. The relation between the amount of time the ligand is in contact with the activated surface and the amount of ligand bound is not linear as the sensor chip reaches saturation.
How much ligand to immobilize?
The amount of ligand to be immobilized depends on the application.
- For specificity measurements, almost any ligand density will do as long as it gives a good signal.
- Concentration measurements need the highest ligand density to facilitate mass transfer limitation. In a total mass transfer controlled experiment, binding will depend on the analyte concentration and not on the binding kinetics between the ligand and analyte.
- Affinity ranking can be done with low to moderate density sensor chips. It is important that the analyte saturates the ligand within a proper time frame.
- Kinetics should be done with the lowest ligand density that still gives a good response without being disturbed by secondary factors such as mass transfer or steric hindrance.
- Low molecular mass binding should be done with high-density sensor chips to bind as much as possible of the analyte to gain proper signal.
In general, for kinetic measurements, a total analyte response of maximal 100 RU, when the analyte is injected (1),(2) is desired (see mass transport). With this value in mind (Rmax), the amount of ligand (in response units) to be immobilized can be calculated with:

Alternatively, more conveniently:
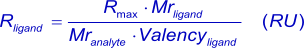
Deactivation
The deactivation solution will block all remaining activated sites with an excess of reagent and because of its high ionic strength and high pH, the solution will wash away most of the electrostatically bound ligand. The amine coupling procedure is usually blocked with ethanolamine, but the use of BSA or casein is also possible. If high salt concentrations are detrimental to the ligand, the experimenter can just wait until all active sites are decayed back to carboxylic groups. The goal of the blocking is to remove the activated groups and make the surface as inert as possible.
In cases where positively charged analytes are being analysed, the surface can be blocked with ethylenediamine to reduce the negative charge of the sensor surface and thus decrease the potential for non-specific binding.
References
| (1) | Karlsson, R. et al Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. Journal of Immunological Methods 229-240; (1991). |
| (2) | Myszka, D. G. Survey of the 1998 optical biosensor literature. J.Mol.Recognit. 12: 390-408; (1999). Goto reference |