This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Issue with my blocking step??
- raulu95
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 8 months ago #1
by raulu95
Issue with my blocking step?? was created by raulu95
I am currently working with a protein-protein interaction that has been a pain, mainly due to the fact that my analyte is incredibly sticky to the reference channel. I use a carboxyl sensor chip and have been doing these experiments on an OpenSPR (Nicoya) which is less sensitive I believe than the Biacore. My ligand has a pH of about 6 while my analyte has a pH of about 9 so I went with the more neutral ethylenediamine as my blocking agent over ethanolamine (is this reasonable?). These are some steps I have taken:
1. BSA blocking at 1 mg/mL (for ref. channel) followed by ethylenediamine blocking. Saw nonspecific binding to sensor chip (signal observed for both channels).
2. Increased BSA concentration and included BSA in running buffer. Same result.
3. Lysozyme blocking at 2 mg/mL (for ref. channel) followed by ethylenediamine blocking. Then switched to buffer containing 1 mg/mL BSA
4. Only ethylenediamine blocking
I suppose my next step would be to try incorporating CMD into the running buffer but would you have any suggestions on things I can do to improve NSB (I seemed to have tried everything typically recommended).
One other thing. I noticed an odd looking phenomenon happening after injecting ethylenediamine. Basically the signals from both channels decrease quite significantly (the picture I am showing here is from experiment 4 where I only blocked with ethylenediamine). Is there something wrong with how I prepare it? I tried experiments with 1 M ethylenediamine in water and in 50 mM sodium borate buffer (adjusted to pH 8.5). I always filter this solution as well. It almost seems like the ethylenediamine is stripping off some of my ligand. Do you think this is happening and, if so, what would you recommend doing?
1. BSA blocking at 1 mg/mL (for ref. channel) followed by ethylenediamine blocking. Saw nonspecific binding to sensor chip (signal observed for both channels).
2. Increased BSA concentration and included BSA in running buffer. Same result.
3. Lysozyme blocking at 2 mg/mL (for ref. channel) followed by ethylenediamine blocking. Then switched to buffer containing 1 mg/mL BSA
4. Only ethylenediamine blocking
I suppose my next step would be to try incorporating CMD into the running buffer but would you have any suggestions on things I can do to improve NSB (I seemed to have tried everything typically recommended).
One other thing. I noticed an odd looking phenomenon happening after injecting ethylenediamine. Basically the signals from both channels decrease quite significantly (the picture I am showing here is from experiment 4 where I only blocked with ethylenediamine). Is there something wrong with how I prepare it? I tried experiments with 1 M ethylenediamine in water and in 50 mM sodium borate buffer (adjusted to pH 8.5). I always filter this solution as well. It almost seems like the ethylenediamine is stripping off some of my ligand. Do you think this is happening and, if so, what would you recommend doing?
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 8 months ago #2
by Arnoud
Replied by Arnoud on topic Issue with my blocking step??
Hi,
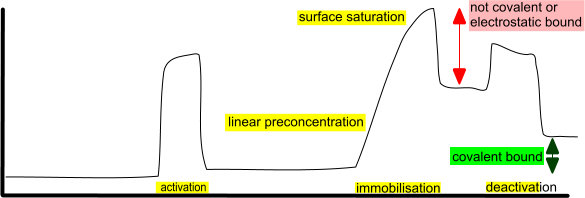
I looked at your sensorgram and made the cartoon below for clarification.
The pre-concentration is linear because it is under full mass transport which is to be expected. The pre-concentration is very efficient and even leads to the saturation of the sensor surface. As can be seen, pre-concentration is not the same as making covalent bonds since after the ligand injection the response level drops significantly. When the ligand injection stops all loosely bound ligand is washed away and covalent and strong electrostatic ligand remains on the chip until the deactivation solution is used. The deactivation solution inactivates and removes the electrostatic bound ligand due to the high pH and high concentration. This gives the impression that the deactivation strips the ligand from the sensor but it was never covalently bound. Still you have 1000 response units.
The response-loss in the reference channel looks as if the NHS/EDC activation is replaced by the ethylene diamine which is smaller. In the end the reference goes back to baseline as before activation.
On solution to this is to use shorter ligand pulses and a lower ligand concentration during the immobilisation. The aim is that after the ligand pulse stops there is not much lowering in response (most ligand is covalently bound we hope).
To overcome the static interaction you can add some salt in the buffer but this will also lower the pre-concentration effect since it will mask charges.
The best way to overcome the analyte non-specific interaction is probably to switch to another type of sensor surface when possible. It is possible to even make your own.
1. Drake, A. W., Tang, M. L., Papalia, G. A., et al.; Biacore surface matrix effects on the binding kinetics and affinity of an antigen/antibody complex. Analytical Biochemistry (429) 1: 58-69; 2012.
2. Masson, J. F., Battaglia, T. M., Davidson, M. J., et al.; Biocompatible polymers for antibody support on gold surfaces. Talanta (67) 5: 918-925; 2005.
3. Masson, J. F., Battaglia, T. M., Kim, Y. C., et al.; Preparation of analyte-sensitive polymeric supports for biochemical sensors. Talanta (64) 3: 716-725; 2004.
4. Rodriguez Emmenegger, C., Brynda, E., Riedel, T., et al.; Interaction of Blood Plasma with Antifouling Surfaces. Langmuir (25) 11: 6328-33; 2000.
I looked at your sensorgram and made the cartoon below for clarification.
The pre-concentration is linear because it is under full mass transport which is to be expected. The pre-concentration is very efficient and even leads to the saturation of the sensor surface. As can be seen, pre-concentration is not the same as making covalent bonds since after the ligand injection the response level drops significantly. When the ligand injection stops all loosely bound ligand is washed away and covalent and strong electrostatic ligand remains on the chip until the deactivation solution is used. The deactivation solution inactivates and removes the electrostatic bound ligand due to the high pH and high concentration. This gives the impression that the deactivation strips the ligand from the sensor but it was never covalently bound. Still you have 1000 response units.
The response-loss in the reference channel looks as if the NHS/EDC activation is replaced by the ethylene diamine which is smaller. In the end the reference goes back to baseline as before activation.
On solution to this is to use shorter ligand pulses and a lower ligand concentration during the immobilisation. The aim is that after the ligand pulse stops there is not much lowering in response (most ligand is covalently bound we hope).
To overcome the static interaction you can add some salt in the buffer but this will also lower the pre-concentration effect since it will mask charges.
The best way to overcome the analyte non-specific interaction is probably to switch to another type of sensor surface when possible. It is possible to even make your own.
1. Drake, A. W., Tang, M. L., Papalia, G. A., et al.; Biacore surface matrix effects on the binding kinetics and affinity of an antigen/antibody complex. Analytical Biochemistry (429) 1: 58-69; 2012.
2. Masson, J. F., Battaglia, T. M., Davidson, M. J., et al.; Biocompatible polymers for antibody support on gold surfaces. Talanta (67) 5: 918-925; 2005.
3. Masson, J. F., Battaglia, T. M., Kim, Y. C., et al.; Preparation of analyte-sensitive polymeric supports for biochemical sensors. Talanta (64) 3: 716-725; 2004.
4. Rodriguez Emmenegger, C., Brynda, E., Riedel, T., et al.; Interaction of Blood Plasma with Antifouling Surfaces. Langmuir (25) 11: 6328-33; 2000.
Please Log in or Create an account to join the conversation.
- raulu95
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 8 months ago #3
by raulu95
Replied by raulu95 on topic Issue with my blocking step??
Thanks Arnoud! I will definitely read those resources shortly. One thing I did want to clarify though- why is it that a smaller ligand pulse (lower interaction time?) and concentration would help with covalent binding efficiency? I always assumed that having a large interaction time with greater concentration would help with getting a sufficient immobilization.
Also, I currently use an interaction time of 500 s and 25 ug/mL ligand. I realize every instrument is different but what would you propose trying if I would need to alter these conditions?
Also, I currently use an interaction time of 500 s and 25 ug/mL ligand. I realize every instrument is different but what would you propose trying if I would need to alter these conditions?
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 8 months ago - 4 years 8 months ago #4
by Arnoud
Replied by Arnoud on topic Issue with my blocking step??
Your main question was if the deactivation was stripping the ligand from the sensor surface. Since the pre-concentration was a kind of overloadig the surface it looks like that the deactivation was doing that. By using a lower ligand concentration during immobilisation you can better predict what the the amount of immobilized ligand will be after deactivation.
With a lower ligand concentration it will take maybe a little longer (it goes slower) to get a high immobilisation level but it will probably conserve ligand as well.
Sufficient immobilization depends on the ligand-analyte size ratio and the purpose of the experiments. I suggest reading www.sprpages.nl/immobilization/immobilization-strategy and other related pages.
Arnoud
With a lower ligand concentration it will take maybe a little longer (it goes slower) to get a high immobilisation level but it will probably conserve ligand as well.
Sufficient immobilization depends on the ligand-analyte size ratio and the purpose of the experiments. I suggest reading www.sprpages.nl/immobilization/immobilization-strategy and other related pages.
Arnoud
Last edit: 4 years 8 months ago by Arnoud.
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud