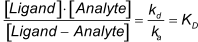This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Ka and Kd results
- Bigsby
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
5 years 6 months ago #1
by Bigsby
Ka and Kd results was created by Bigsby
I'm new to SPR having just finished my first experiment and I'm still not quite clear on interpreting some of the kinetics results, specifically Kd. Also I'm trying to understand how to translate Ka and Kd into how fast my analyte binds to the ligand and how fast it unbinds - so as to determine hypothetical viability of my analyte as a drug, for example if I was testing a pain medication it would be desirable for it to bind quickly and then take a long time to unbind.
I understand Ka represents amount of interactions in 1 M of solution, per second. But I'm still not quite understanding Kd after reading the description on this site. And how would those values (Ka and Kd) translate into whether the analyte binds faster or slower then it unbinds?
So my results were as such - ka = 9.458E+4, kd = 9.787E-4 and KD = 1.035E-8
I know this is probably basic stuff for some of the users on this site but for me it's all very new so any help in understating this further would be greatly appreciated.
I understand Ka represents amount of interactions in 1 M of solution, per second. But I'm still not quite understanding Kd after reading the description on this site. And how would those values (Ka and Kd) translate into whether the analyte binds faster or slower then it unbinds?
So my results were as such - ka = 9.458E+4, kd = 9.787E-4 and KD = 1.035E-8
I know this is probably basic stuff for some of the users on this site but for me it's all very new so any help in understating this further would be greatly appreciated.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
5 years 6 months ago - 5 years 6 months ago #2
by Arnoud
Replied by Arnoud on topic Ka and Kd results
Hi and welcome,
There is quite a confusion in your parameter notation, so first of all, let's get the parameters right.
The association and dissociation rate constants always in lower case en the equilibrium dissociation constants always in upper case. The relation between the association and dissociation is:
The equilibrium dissociation constant is a measure of the overall strength of the interaction. When it is lower, the binding is stronger. As can be seen from the formula you can have a low KD either by a low kd or a high ka (or both). You can visualize this with an affinity plot ( www.sprpages.nl/sensorgram-tutorial/affinity-plot ).
If the compounds dissociates faster than it binds you would have no binding .
.
Depending on the application a fast association or slow dissociation is preferred. The association rate can be influenced by the compound concentration (ka in M-1s-1) but the dissociation rate not (kd in s-1).
Kinds regards
Arnoud
There is quite a confusion in your parameter notation, so first of all, let's get the parameters right.
| Parameter | Meaning | Units |
| ka | Association rate constant | M-1s-1 |
| kd | Dissociation rate constant | s-1 |
| KD | Equilibrium dissociation constant | M |
The association and dissociation rate constants always in lower case en the equilibrium dissociation constants always in upper case. The relation between the association and dissociation is:
The equilibrium dissociation constant is a measure of the overall strength of the interaction. When it is lower, the binding is stronger. As can be seen from the formula you can have a low KD either by a low kd or a high ka (or both). You can visualize this with an affinity plot ( www.sprpages.nl/sensorgram-tutorial/affinity-plot ).
If the compounds dissociates faster than it binds you would have no binding
Depending on the application a fast association or slow dissociation is preferred. The association rate can be influenced by the compound concentration (ka in M-1s-1) but the dissociation rate not (kd in s-1).
Kinds regards
Arnoud
Last edit: 5 years 6 months ago by Arnoud.
Please Log in or Create an account to join the conversation.
- Bigsby
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
5 years 5 months ago - 5 years 5 months ago #3
by Bigsby
Replied by Bigsby on topic Ka and Kd results
Ok thank you for the explanation. So just to be clear then for an example, in my results where ka = 9.458E+4 and kd = 9.787E-4. This means that I am getting 9.458E+4 association events per mole of analyte per second. And that I am getting 9.787E-4 disassociation events per second (no influence by concentration)
Which then implies that one my analyte binds, as ka is higher then kd, but also that my analyte is binding faster than it is unbinding, as the number for association is much higher than disassociation, per second.
Is that correct?
Further to this, is there a known cut-off value for ka and kd for either to be determined fast or slow in regards to association and dissociation?
Many thanks,
Bigsby
Which then implies that one my analyte binds, as ka is higher then kd, but also that my analyte is binding faster than it is unbinding, as the number for association is much higher than disassociation, per second.
Is that correct?
Further to this, is there a known cut-off value for ka and kd for either to be determined fast or slow in regards to association and dissociation?
Many thanks,
Bigsby
Last edit: 5 years 5 months ago by Bigsby.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
5 years 5 months ago #4
by Arnoud
Replied by Arnoud on topic Ka and Kd results
At first glance you can say that (maybe / maybe not). The association phase is not only association; it is also dissociation, equilibrium and saturation. Immediately as a compounds binds it starts to dissociate. At start, the amount of associations will be larger than the amount of dissociations and you have effectively a binding. But as binding progresses more compound is bound and more dissociations occur until the point that there are as many binding events as unbinding events. This situation is called steady state (equilibrium). At this point the association rate is as fast as the dissociation rate.
Remember the formula two posts back about the concentrations of the ligand, analyte and complex. When binding analyte to the ligand, the concentration free ligand is getting lower and the concentration complex is getting higher. Therefore you get a shift from almost full association rate to equilibrium. Removing the analyte leads to a full dissociation provided that the analyte is washed away fast enough and is not rebinding.
The association and dissociation rate constants are parameters to model association and dissociation rate. And because the underlying kinetics is different (as can be seen in the units) it is in my opinion not prudent to compare ka and kd like you do.
I am not aware of established cut-off values for ka and kd. Dissociation rates below 10-4 s-1 are considered relative slow but for high affinity you may want to have 10-5 s-1.
Arnoud
Remember the formula two posts back about the concentrations of the ligand, analyte and complex. When binding analyte to the ligand, the concentration free ligand is getting lower and the concentration complex is getting higher. Therefore you get a shift from almost full association rate to equilibrium. Removing the analyte leads to a full dissociation provided that the analyte is washed away fast enough and is not rebinding.
The association and dissociation rate constants are parameters to model association and dissociation rate. And because the underlying kinetics is different (as can be seen in the units) it is in my opinion not prudent to compare ka and kd like you do.
I am not aware of established cut-off values for ka and kd. Dissociation rates below 10-4 s-1 are considered relative slow but for high affinity you may want to have 10-5 s-1.
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud