This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
low KD but early saturation
- uptracy
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 10 months ago #1
by uptracy
low KD but early saturation was created by uptracy
I am studying a protein-ligand interaction. The ligand is fixed on a sensorchip with protein flowing through the channel. The binding affinity (KD) of the wild-type protein and the mutant were compared. The wt protein showed a stronger signal intensity than the mutant at the same concentration. While within a series of concentrations, the wt reached saturation at a higher concentration. But its KD obtained from the steady-state affinity is smaller than the mutant. If I understand correctly, the higher affinity molecule should reach saturation earlier than the weaker binding molecule. So what might be the reason to explain this contradiction?
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 10 months ago - 4 years 10 months ago #2
by Arnoud
Replied by Arnoud on topic low KD but early saturation
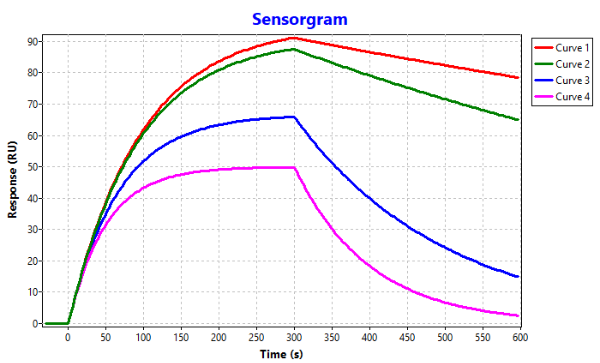
Practically you can't say that since the time to steady state depends on the ka, kd and analyte concentration. As can be seen in the simulation the red line has the highest affinity. But due to the slow kd it takes 285 seconds (tReq) to reach 95% of the theoretical Req. While the curve with the lowest affinity has the fastest kd it will reach steady state in 150 seconds at the same analyte concentration.
However, the high affinity interaction reaches more response than the low affinity interaction and this is because of the different analyte concentrations ratio compared to the KD.
Even when the analyte injection is very long the Req response wil be different (Req column).
For more on this topic download this PDF " www.sprpages.nl/component/phocadownload/...ad=23:forum-affinity ".
Arnoud
However, the high affinity interaction reaches more response than the low affinity interaction and this is because of the different analyte concentrations ratio compared to the KD.
Even when the analyte injection is very long the Req response wil be different (Req column).
For more on this topic download this PDF " www.sprpages.nl/component/phocadownload/...ad=23:forum-affinity ".
Arnoud
Last edit: 4 years 10 months ago by Arnoud. Reason: typos
Please Log in or Create an account to join the conversation.
- uptracy
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 10 months ago #3
by uptracy
Replied by uptracy on topic lower affinity but lower concentration needed to saturate the ligand
Thank you for your explanation. But I think I did not clarify my question. The saturation I meant is not the steady state. I meant the ligand saturation. I was testing the binding affinity of WT and mutants (M1 and M2) with the ligand in the range of 1,10,20,30,40,50,60,70,80 uM. The wt had a stronger signal than the mutant1 (M1) at each concentration. From 1 to 80 uM, I saw a gradually increasing signal at the steady-state. While M1 seems saturated the ligand at 60 uM already as there is no big difference among 60,70 and 80 uM in terms of signal intensity. My understanding is that M1 has a lower affinity, so it should need more concentration to get the ligand saturated than the WT. But the experiment showed the opposite. However, for the other mutant M2, it had a lower affinity than the WT and it did show far from the saturation at the same concentration with the WT. So any suggestions to fix this odd?
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 10 months ago #4
by Arnoud
Replied by Arnoud on topic lower affinity but lower concentration needed to saturate the ligand
OK I get confused about how you call things. Saturation is the moment that all the ligand is occupied by the analyte and steady state when the amount of associations are the same as the amount of dissociations. Where saturation can be at steady state, steady state is almost never saturating the ligand (
www.sprpages.nl/sensorgram-tutorial/a-curve
).
Do the mutants differ only in some amino acids and not in glycosilation (mass)?
A major error here can be the concentration determination of the mutants vs WT.
The next thing is that for steady state analysis (equilibrium analysis) all curves must be at steady state. Please show the sensorgrams.
And one concern about the figure you show. I tried a steady state fitting on the values of your graph but the fit was not well. The 1 µM concentration has already a major response for Wt and M1. Are you sure you looking at specific binding? Again, please show the original sensorgrams.
Kind regards
Arnoud
Do the mutants differ only in some amino acids and not in glycosilation (mass)?
A major error here can be the concentration determination of the mutants vs WT.
The next thing is that for steady state analysis (equilibrium analysis) all curves must be at steady state. Please show the sensorgrams.
And one concern about the figure you show. I tried a steady state fitting on the values of your graph but the fit was not well. The 1 µM concentration has already a major response for Wt and M1. Are you sure you looking at specific binding? Again, please show the original sensorgrams.
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- uptracy
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
4 years 10 months ago #5
by uptracy
Replied by uptracy on topic lower affinity but lower concentration needed to saturate the ligand
Sorry for the confusion. And there were mistakes in the value of the steady-state in my last figure. And here I presented the correct one and three sensorgrams for WT, M1 and M2. The concentration is bottom-up, 1-80 uM. The three proteins only differ in one amino acid without any glycosylation. At this point, I believe the concentrations are right for all. Also, I have other experiments show it is a specific binding. So I really got confused by these results.
Much appreciated.
Much appreciated.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
4 years 10 months ago #6
by Arnoud
Replied by Arnoud on topic lower affinity but lower concentration needed to saturate the ligand
Looking at the sensorgrams I noticed that the curves are fairly equally spaced when injecting the higher analyte concentrations. I would expect that the highest 2-3 concentrations should be closer to each other indication a true saturation of the ligand. Now it looks like that there is some secundairy binding which keeps increasing at higher concentrations.
Maybe this is also the explanation of the difference in response between WT and mutant.
(Note: I am not used to this small step titrations. In general it is easier to make a step 2 or step 3 dilution. And this will give the same information and result.)
Therefore I think that the problem of the saturation and/or reversed affinity is an artefact of the interaction system.
Arnoud
Maybe this is also the explanation of the difference in response between WT and mutant.
(Note: I am not used to this small step titrations. In general it is easier to make a step 2 or step 3 dilution. And this will give the same information and result.)
Therefore I think that the problem of the saturation and/or reversed affinity is an artefact of the interaction system.
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud