This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
ag-ab reactions, what is the different between capture assay and direct assay?
- taoguo
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
5 years 11 months ago #1
by taoguo
ag-ab reactions, what is the different between capture assay and direct assay? was created by taoguo
Hi,
I am working on antigen-antibody reactions by SPR. and the data is inconsistent if we measure the KD value by different ways. what is the different between them and why?
method 1, immobilized antigen and then injected ab (5 concentrations, regenerate the chip each time)
method 2, immobilized anti-human IgG, and captured ab, and then injected ag (5 concentrations, regenerate the chip each time).
the data was fit to the equation for Langmuir 1:1 model.
thanks in advance!
gt
I am working on antigen-antibody reactions by SPR. and the data is inconsistent if we measure the KD value by different ways. what is the different between them and why?
method 1, immobilized antigen and then injected ab (5 concentrations, regenerate the chip each time)
method 2, immobilized anti-human IgG, and captured ab, and then injected ag (5 concentrations, regenerate the chip each time).
the data was fit to the equation for Langmuir 1:1 model.
thanks in advance!
gt
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
5 years 11 months ago #2
by Arnoud
Replied by Arnoud on topic ag-ab reactions, what is the different between capture assay and direct assay?
Dear gt,
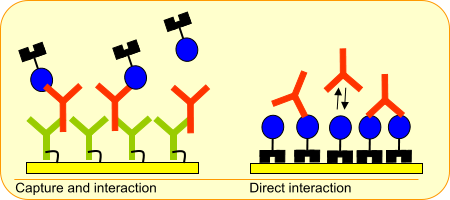
Your method 1 is a direct binding of antibody to the antigen. Since an antibody has two binding sites this can result in a bivalent interaction (1 antibody: 2 antigens). The second method gives a 1:1 interaction because the two binding sites of the antibody work independently here.
You can model the bivalent with the bvalent model, but be aware that the reaction can be mixed as can be seen in the cartoon.
kind regards
Arnoud
Your method 1 is a direct binding of antibody to the antigen. Since an antibody has two binding sites this can result in a bivalent interaction (1 antibody: 2 antigens). The second method gives a 1:1 interaction because the two binding sites of the antibody work independently here.
You can model the bivalent with the bvalent model, but be aware that the reaction can be mixed as can be seen in the cartoon.
kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- taoguo
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
5 years 11 months ago #3
by taoguo
Replied by taoguo on topic ag-ab reactions, what is the different between capture assay and direct assay?
Thank you so much, Arnoud!
So the method 2 gives a more reliable result, right? I am wondering why the two binding sites work independently in this case? if one site is occupied, the other one could not bind to antigen? when we immobilize antibody onto the surface, the two sites still work independently?
For method1, if we lower the amount of ligand (antigen) immobilized on the chip, can we get a 1:1 interaction?
thanks for any comments.
gt
So the method 2 gives a more reliable result, right? I am wondering why the two binding sites work independently in this case? if one site is occupied, the other one could not bind to antigen? when we immobilize antibody onto the surface, the two sites still work independently?
For method1, if we lower the amount of ligand (antigen) immobilized on the chip, can we get a 1:1 interaction?
thanks for any comments.
gt
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
5 years 11 months ago #4
by Arnoud
Replied by Arnoud on topic ag-ab reactions, what is the different between capture assay and direct assay?
Yes the second method gives the best results. This is because each arm of the antibody has the same change of binding an analyte. And if the analyte is small enough, each antibody can bind two analytes in an independent manner.
When the antibody is used as analyte both arms have the same change of binding the ligand, but when one arm is bound, the second arm can only bind a ligand that is close by. The second binding will not add mass to the system and is not detected as such. However, by binding with two arms the overall kinetics will become stronger (the dissociation seems to be slower) since two arms have to dissociate before the antibody can leave.
By lowering the ligand concentration, the overall binding will shift to the more 1:1 model, but you should do many experiments (immobilizations) to prove that you have 100% 1:1 interaction.
1. Tiwari, P. B., Üren, A., He, J., et al.; Note: Model identification and analysis of bivalent analyte surface plasmon resonance data. Review of Scientific Instruments (86) 10: 106107; 2015.
Kind regards
Arnoud
When the antibody is used as analyte both arms have the same change of binding the ligand, but when one arm is bound, the second arm can only bind a ligand that is close by. The second binding will not add mass to the system and is not detected as such. However, by binding with two arms the overall kinetics will become stronger (the dissociation seems to be slower) since two arms have to dissociate before the antibody can leave.
By lowering the ligand concentration, the overall binding will shift to the more 1:1 model, but you should do many experiments (immobilizations) to prove that you have 100% 1:1 interaction.
1. Tiwari, P. B., Üren, A., He, J., et al.; Note: Model identification and analysis of bivalent analyte surface plasmon resonance data. Review of Scientific Instruments (86) 10: 106107; 2015.
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud