This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Is this similar to kinetic titration, right?
- Koreanraichu
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
7 years 7 months ago #1
by Koreanraichu
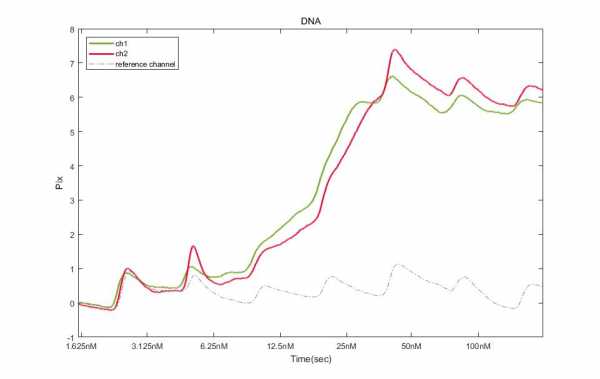
(dotted line : no bind DNA, reference channel/line : bind DNA)
I injected 10min and no washing. start point is 1.625nM and end point is 100nM, reference channel shows peak(like zig-zag). but DNA rise up when 6.25nM and 12.5nM. Is that rise up because concentration is saturated? or do I extend inject time?
Is this similar to kinetic titration, right? was created by Koreanraichu
(dotted line : no bind DNA, reference channel/line : bind DNA)
I injected 10min and no washing. start point is 1.625nM and end point is 100nM, reference channel shows peak(like zig-zag). but DNA rise up when 6.25nM and 12.5nM. Is that rise up because concentration is saturated? or do I extend inject time?
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
7 years 7 months ago #2
by Arnoud
Replied by Arnoud on topic Is this similar to kinetic titration, right?
Hi,
The term kinetic titration and single cycle kinetics are both used for the same thing: subsequent higher analyte injection with or without dissociation in between.
I think you see some binding starting at the concentration of 3 nM up to 25 nM. I have no experience with DNA binding, but the interaction looks stable.
Probably you have saturated the ligand on the sensor chip.
How did you make your analyte dilutions? Did you make the 100 nM and made a serial dilution to 50, 25 etc? I ask this because you expect the zigzag of the no binidng DNA to get higher with higher DNA concnetrations and the 50 and 100 seems to be lower.
Kind regards
Arnoud
The term kinetic titration and single cycle kinetics are both used for the same thing: subsequent higher analyte injection with or without dissociation in between.
I think you see some binding starting at the concentration of 3 nM up to 25 nM. I have no experience with DNA binding, but the interaction looks stable.
Probably you have saturated the ligand on the sensor chip.
How did you make your analyte dilutions? Did you make the 100 nM and made a serial dilution to 50, 25 etc? I ask this because you expect the zigzag of the no binidng DNA to get higher with higher DNA concnetrations and the 50 and 100 seems to be lower.
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- Koreanraichu
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
7 years 7 months ago #3
by Koreanraichu
Replied by Koreanraichu on topic Is this similar to kinetic titration, right?
How did you make your analyte dilutions? Did you make the 100 nM and made a serial dilution to 50, 25 etc? I ask this because you expect the zigzag of the no binidng DNA to get higher with higher DNA concnetrations and the 50 and 100 seems to be lower.
-->Oh, yes. I diluted 2x. but injection rate is 40ul/min and 10min, 3tube inject, so I started at 1.5ml/200nM DNA for 100nM~1.625nM(diluted times).
-->Oh, yes. I diluted 2x. but injection rate is 40ul/min and 10min, 3tube inject, so I started at 1.5ml/200nM DNA for 100nM~1.625nM(diluted times).
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud