This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Sensorgrams - inaccurate fits
- Layzing
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
9 years 8 months ago #1
by Layzing
Sensorgrams - inaccurate fits was created by Layzing
Hi!
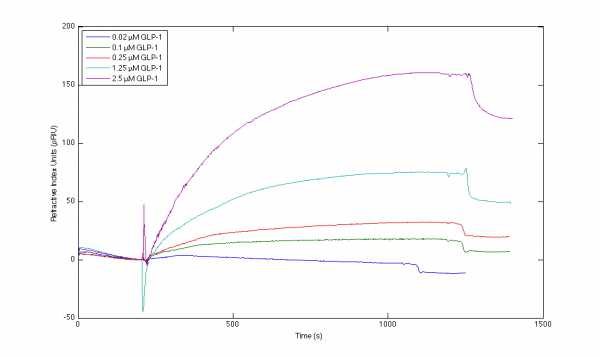
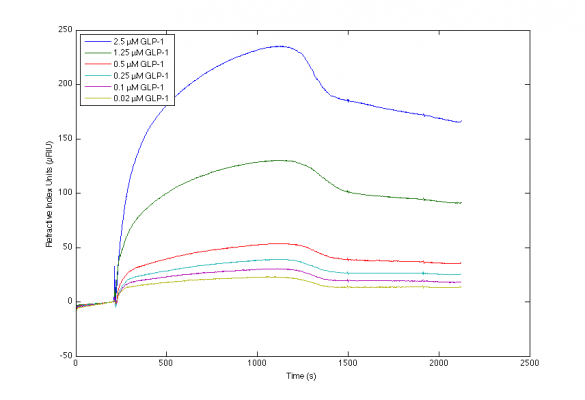
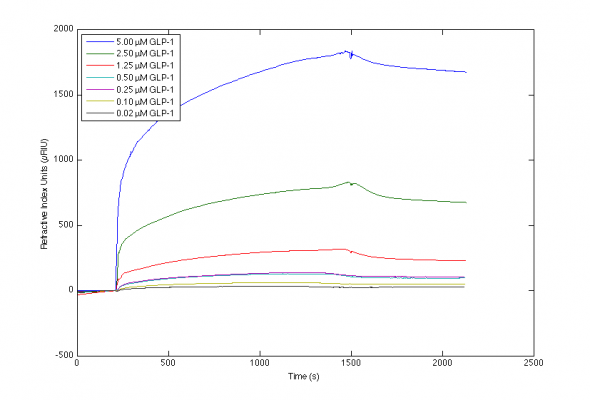
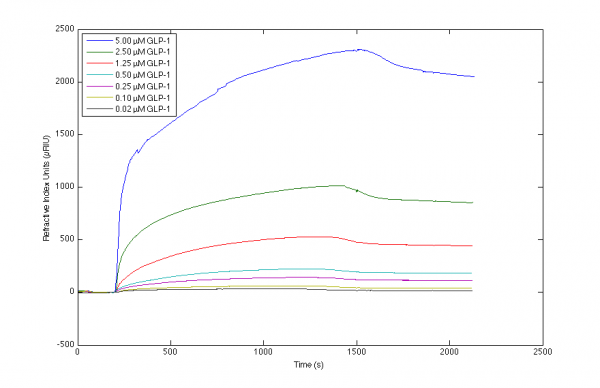
I've been working with the interaction between various antibodies and GLP-1, and achieved the included sensorgrams. I have trouble producing a valid mathematical fit to the curvatures, and although I've tried to locate advice on this page and the included references, I'm still uncertain which parameters to alter to obtain sensorgrams of higher quality.
Please let me know what your more experienced eyes notice about my sensorgrams, and what you would adjust if any.
Settings:
Chip: Xantec carboxymethylated hydrogel (HC) 1000m
Flow rate: 20 uL/min
Running buffer: PBS buffer
Protein solutions: 1 mg/ml GLP-1 in 20 mM AcOH solution diluted into PBS buffer for desired concentrations
Results from capture immobilisation method at 25 C.
Results from direct immobilisation method at 25 C.
Results from direct immobilisation method at 10 C.
Results from direct immobilisation method at 40 C.
I've been working with the interaction between various antibodies and GLP-1, and achieved the included sensorgrams. I have trouble producing a valid mathematical fit to the curvatures, and although I've tried to locate advice on this page and the included references, I'm still uncertain which parameters to alter to obtain sensorgrams of higher quality.
Please let me know what your more experienced eyes notice about my sensorgrams, and what you would adjust if any.
Settings:
Chip: Xantec carboxymethylated hydrogel (HC) 1000m
Flow rate: 20 uL/min
Running buffer: PBS buffer
Protein solutions: 1 mg/ml GLP-1 in 20 mM AcOH solution diluted into PBS buffer for desired concentrations
Results from capture immobilisation method at 25 C.
Results from direct immobilisation method at 25 C.
Results from direct immobilisation method at 10 C.
Results from direct immobilisation method at 40 C.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
9 years 8 months ago - 9 years 8 months ago #2
by Arnoud
Replied by Arnoud on topic Sensorgrams - inaccurate fits
Hi,
Just looking at the sensorgrams, I have the following remarks:
Sensorgram 1) I do not see any dissociation part and the blue line looks a to have a shorter injection period. In addition it looks like that there was some negative drift.
2) The transition of the association to the dissociation phase is strange as if the separation of the sample and buffer was not optimal.
3) Too high response levels and probably mass transfer at the beginning of the sensorgram.
4) As 3) but highest is more bumpy and the transition of association to dissociation phase is diffrent from the rest of the dissociation.
Suggestions
- keep responses low
- equilibrate thoroughly
- have sufficient dissociation
- check separation of sample and buffer (clean tubing)
I think with some optimization you can have nice data
Kind regards
Arnoud
Just looking at the sensorgrams, I have the following remarks:
Sensorgram 1) I do not see any dissociation part and the blue line looks a to have a shorter injection period. In addition it looks like that there was some negative drift.
2) The transition of the association to the dissociation phase is strange as if the separation of the sample and buffer was not optimal.
3) Too high response levels and probably mass transfer at the beginning of the sensorgram.
4) As 3) but highest is more bumpy and the transition of association to dissociation phase is diffrent from the rest of the dissociation.
Suggestions
- keep responses low
- equilibrate thoroughly
- have sufficient dissociation
- check separation of sample and buffer (clean tubing)
I think with some optimization you can have nice data
Kind regards
Arnoud
Last edit: 9 years 8 months ago by Arnoud.
Please Log in or Create an account to join the conversation.
- Layzing
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
9 years 8 months ago - 9 years 8 months ago #3
by Layzing
Replied by Layzing on topic Sensorgrams - inaccurate fits
Thank you very much for your quick reply.
I've discovered a more precise method to separate the sample and buffer when injecting. Hopefully this will improve the transition, especially regarding the ones immobilised direct.
Additional quick question regarding #1. Do you know what the quick drop after association is related to? I've performed additional experiments with indirect immobilisation on another chip, and the case seems to be identical.
I've discovered a more precise method to separate the sample and buffer when injecting. Hopefully this will improve the transition, especially regarding the ones immobilised direct.
Additional quick question regarding #1. Do you know what the quick drop after association is related to? I've performed additional experiments with indirect immobilisation on another chip, and the case seems to be identical.
Last edit: 9 years 8 months ago by Layzing.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
9 years 8 months ago #4
by Arnoud
Replied by Arnoud on topic Sensorgrams - inaccurate fits
First i was thinking about a buffer difference between flow and analyte but since it is not obvious present at the start probably that is not the case.
Speculation: Maybe a built up of protein that binds very weak and is washed away immediately after stopping the analyte injection. After that the dissociation of the remaining thighter bound protein is visible.
Arnoud
Speculation: Maybe a built up of protein that binds very weak and is washed away immediately after stopping the analyte injection. After that the dissociation of the remaining thighter bound protein is visible.
Arnoud
Please Log in or Create an account to join the conversation.
- Layzing
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
9 years 7 months ago #5
by Layzing
Replied by Layzing on topic Sensorgrams - inaccurate fits
Is it ever reasonable to treat such data in steps, circumventing the sudden non-BI related jump?
Calculating dissocation constant isolated and keeping it constant for the association fit?
Calculating dissocation constant isolated and keeping it constant for the association fit?
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
9 years 7 months ago #6
by Arnoud
Replied by Arnoud on topic Sensorgrams - inaccurate fits
It is possible to do that but I have never seen anyone publishing it.
One other concern is that you can pick the region to be fit at your convenience. That will give some bias to the answer, making it look better than it is.
I suggest you try, but do some simulations with the fitted values and compare the simulations with the original data. In the end you should show a sensorgram and fitting to your audience.
Arnoud
One other concern is that you can pick the region to be fit at your convenience. That will give some bias to the answer, making it look better than it is.
I suggest you try, but do some simulations with the fitted values and compare the simulations with the original data. In the end you should show a sensorgram and fitting to your audience.
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud