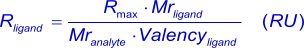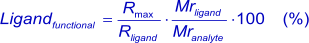This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Determination of protein concentration on the chip
- asingh
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
10 years 3 weeks ago #1
by asingh
Determination of protein concentration on the chip was created by asingh
To Admins of the website
The link does not work at
www.sprpages.nl/downloads/how-to.html
and user gets following error
View not found [name, type, prefix]: category, pdf, contentView
Question in general:
How to determine the concentration of immobilized ligand on the chip ? is there a direct formula ?
Thanks
Aman
The link does not work at
www.sprpages.nl/downloads/how-to.html
and user gets following error
View not found [name, type, prefix]: category, pdf, contentView
Question in general:
How to determine the concentration of immobilized ligand on the chip ? is there a direct formula ?
Thanks
Aman
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
10 years 3 weeks ago - 10 years 3 weeks ago #2
by Arnoud
Replied by Arnoud on topic Determination of protein concentration on the chip
@asingh: As far as we know the page is normally accessible. You may wish to refresh your browser since the menu structure has changed.
Do you mean the concentration in pg/mm2?
As a approximation: 1 RU ~ 1 pg/mm2 for a CM5 sensor chip.
However, a CM5 has a thickness of 100 nm so it is not a flat surface.
For the kinetics it is not of interrest how much you immobilize as long as you have a decent signal without mass transport.
Arnoud
Do you mean the concentration in pg/mm2?
As a approximation: 1 RU ~ 1 pg/mm2 for a CM5 sensor chip.
However, a CM5 has a thickness of 100 nm so it is not a flat surface.
For the kinetics it is not of interrest how much you immobilize as long as you have a decent signal without mass transport.
Arnoud
Last edit: 10 years 3 weeks ago by Arnoud.
Please Log in or Create an account to join the conversation.
- asingh
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
10 years 2 weeks ago - 10 years 2 weeks ago #3
by asingh
Thank you Arnoud,
1. Please see the error message I get regardless of browser type (Mozilla, Chrome, IE). All browsers are up to date. I can’t see the pdf.
2. I meant protein concentration in mg/mL or uM . I am sure there is a formula to convert 1 pg/mm2 to mg/mL. The chip I use have thickness of ~50 nm for the gold layer and additional ~2nm for surface coating. I mostly capture around 5000 RU of the protein for a typical fragment screen. I wonder what would be protein concentration in mg/mL on the chip.
3. Please have a look at example data I have for analyte MW of 707 against ligand MW 22500 as monomer. Using 1: 1 binding model I get a decent KD for the protein A and its close homologue protein B using either fastStep or Onestep gradient injections. However the Rmax observed is about 70% of theoretical Rmax when about 5000 RU of both proteins were capture on Ni NTA chips. Could one conclude that protein is 70 % functional on the chip?
Again thank you very much for your replies.
Aman
Replied by asingh on topic Determination of protein concentration on the chip
Thank you Arnoud,
1. Please see the error message I get regardless of browser type (Mozilla, Chrome, IE). All browsers are up to date. I can’t see the pdf.
2. I meant protein concentration in mg/mL or uM . I am sure there is a formula to convert 1 pg/mm2 to mg/mL. The chip I use have thickness of ~50 nm for the gold layer and additional ~2nm for surface coating. I mostly capture around 5000 RU of the protein for a typical fragment screen. I wonder what would be protein concentration in mg/mL on the chip.
3. Please have a look at example data I have for analyte MW of 707 against ligand MW 22500 as monomer. Using 1: 1 binding model I get a decent KD for the protein A and its close homologue protein B using either fastStep or Onestep gradient injections. However the Rmax observed is about 70% of theoretical Rmax when about 5000 RU of both proteins were capture on Ni NTA chips. Could one conclude that protein is 70 % functional on the chip?
Again thank you very much for your replies.
Aman
Last edit: 10 years 2 weeks ago by asingh.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
10 years 2 weeks ago #4
by Arnoud
Replied by Arnoud on topic Determination of protein concentration on the chip
1. Hi Aman, I understand now, fixed the link.
2. I will come back to this soon.
Arnoud
2. I will come back to this soon.
Arnoud
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
10 years 2 weeks ago - 10 years 2 weeks ago #5
by Arnoud
Replied by Arnoud on topic Determination of protein concentration on the chip
Some calculations
In general, for kinetic measurements, a total analyte response of maximal 100 RU, when the analyte is injected (4),(6) is desired (see mass transport). With this value in mind (Rmax), the amount of ligand (in response units) to be immobilized can be calculated with:
Alternatively, more conveniently:
Because of the linear relation between response and amount of protein immobilized to the sensor surface (7), (8 ), the theoretical number of ligand sites after immobilization can be calculated (3).
This formula is theoretical because it assumes that all immobilized ligand molecules are fully accessible and functional, which is most likely not the case with the standard amine coupling. When the Rmax is known after analyte injection, the percentage of functional ligand (the ligand that can bind the analyte) can be calculated. A low analyte response can be caused by low affinity, an impure ligand or that binding sites have been affected by the immobilization procedure. A too high response can be induced by non-specific binding or analyte aggregates (1).
Because the dextran matrix has a significant extension (in of the order of 100 nm for sensor chip CM5), "surface concentrations" on the sensor chip surface are strictly speaking volume concentrations. The relation between the actual protein 'concentration' and the Response units (RU) for the dextran type of sensor chips (2), (5) is:
1000 RU = 1 ng/mm2 surface concentration
and
1000 RU = 10 mg/ml volume concentration
These relations make it possible to calculate the ligand concentration:
From a kinetic point of view the actual ligand (surface or volume concentration) is not of much use because it is not directly used in the kinetic equations. More or less ligand will affect the Rmax but will not change the kinetics of the system. Also, the calculation of the amount bound analyte does, in my opinion, not add any relevant information to the kinetics.
In your question you have a 50 nm gold layer and a 2 nm of surface coating. The thickness of the gold layer will not contribute to the volume concentration of your ligand.
During capture of the His-Tagged protein you will capture all variant with a His-Tag meaning that also incomplete or degraded protein will be captured. It can be beneficial to have the His-Tag at the C-terminal of the protein to minimize this.
Based on what you show I think that you have indeed a protein mixture that is 70% functional active.
Kind regards
Arnoud
References
(1) Biacore AB BIA Symposium '98.
(2) Biacore AB BIACORE Getting Started. (1998).
(3) Biacore AB BIACORE Technology Handbook. (1998).
(4) Karlsson, R., A. Michaelson and L. Mattson Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. Journal of Immunological Methods 229-240; (1991). www.sciencedirect.com/science/article/pii/0022175991903319
(5) Muller, K. M., K. M. Arndt and A. Pluckthun Model and simulation of multivalent binding to fixed ligands. Analytical Biochemistry 261: 149-158; (1998).
(6) Myszka, D. G. Survey of the 1998 optical biosensor literature. J.Mol.Recognit. 12: 390-408; (1999).
onlinelibrary.wiley.com/doi/10.1002/(SIC...-1352(199911/12)12:6 <390::AID-JMR482>3.0.CO;2-8/abstract"
(7) Quinn, J. G., S. O'Neill, A. Doyle, et al. Development and application of surface plasmon resonance-based biosensors for the detection of cell-ligand interactions. Analytical Biochemistry 281: 135-143; (2000). " www.sciencedirect.com/science/article/pii/S0003269700945640 "
(8 ) Stenberg, E., B. Persson, H. Roos, et al. Quantitative determination of surface concentration of protein with surface plasmon resonance by using radiolabelled proteins. (1991).
In general, for kinetic measurements, a total analyte response of maximal 100 RU, when the analyte is injected (4),(6) is desired (see mass transport). With this value in mind (Rmax), the amount of ligand (in response units) to be immobilized can be calculated with:
Alternatively, more conveniently:
Because of the linear relation between response and amount of protein immobilized to the sensor surface (7), (8 ), the theoretical number of ligand sites after immobilization can be calculated (3).
This formula is theoretical because it assumes that all immobilized ligand molecules are fully accessible and functional, which is most likely not the case with the standard amine coupling. When the Rmax is known after analyte injection, the percentage of functional ligand (the ligand that can bind the analyte) can be calculated. A low analyte response can be caused by low affinity, an impure ligand or that binding sites have been affected by the immobilization procedure. A too high response can be induced by non-specific binding or analyte aggregates (1).
Because the dextran matrix has a significant extension (in of the order of 100 nm for sensor chip CM5), "surface concentrations" on the sensor chip surface are strictly speaking volume concentrations. The relation between the actual protein 'concentration' and the Response units (RU) for the dextran type of sensor chips (2), (5) is:
1000 RU = 1 ng/mm2 surface concentration
and
1000 RU = 10 mg/ml volume concentration
These relations make it possible to calculate the ligand concentration:
From a kinetic point of view the actual ligand (surface or volume concentration) is not of much use because it is not directly used in the kinetic equations. More or less ligand will affect the Rmax but will not change the kinetics of the system. Also, the calculation of the amount bound analyte does, in my opinion, not add any relevant information to the kinetics.
In your question you have a 50 nm gold layer and a 2 nm of surface coating. The thickness of the gold layer will not contribute to the volume concentration of your ligand.
During capture of the His-Tagged protein you will capture all variant with a His-Tag meaning that also incomplete or degraded protein will be captured. It can be beneficial to have the His-Tag at the C-terminal of the protein to minimize this.
Based on what you show I think that you have indeed a protein mixture that is 70% functional active.
Kind regards
Arnoud
References
(1) Biacore AB BIA Symposium '98.
(2) Biacore AB BIACORE Getting Started. (1998).
(3) Biacore AB BIACORE Technology Handbook. (1998).
(4) Karlsson, R., A. Michaelson and L. Mattson Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. Journal of Immunological Methods 229-240; (1991). www.sciencedirect.com/science/article/pii/0022175991903319
(5) Muller, K. M., K. M. Arndt and A. Pluckthun Model and simulation of multivalent binding to fixed ligands. Analytical Biochemistry 261: 149-158; (1998).
(6) Myszka, D. G. Survey of the 1998 optical biosensor literature. J.Mol.Recognit. 12: 390-408; (1999).
onlinelibrary.wiley.com/doi/10.1002/(SIC...-1352(199911/12)12:6 <390::AID-JMR482>3.0.CO;2-8/abstract"
(7) Quinn, J. G., S. O'Neill, A. Doyle, et al. Development and application of surface plasmon resonance-based biosensors for the detection of cell-ligand interactions. Analytical Biochemistry 281: 135-143; (2000). " www.sciencedirect.com/science/article/pii/S0003269700945640 "
(8 ) Stenberg, E., B. Persson, H. Roos, et al. Quantitative determination of surface concentration of protein with surface plasmon resonance by using radiolabelled proteins. (1991).
Last edit: 10 years 2 weeks ago by Arnoud.
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud