This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Odd kinetic trace
- lgarner
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
10 years 8 months ago #1
by lgarner
Odd kinetic trace was created by lgarner
Hope you can help!
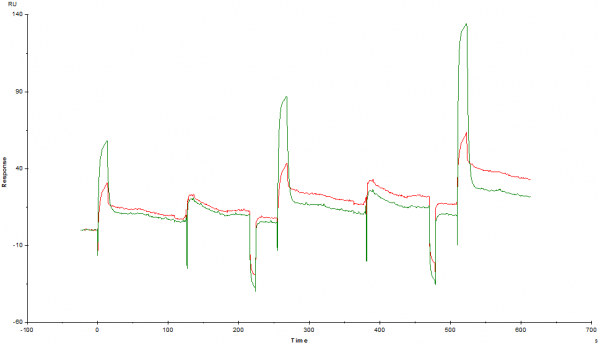
BIAcore 3000, CM5 chip, amine-coupled streptavidin, biotinylated ligands.
FC1 7310 strep + 140 control protein (45kDa)
FC2 6753 strep + 302 ligand protein (75kDa)
100 ul/min, KINJECT 25ul, 90s dissociation
single flow cell injections at maximal data rate (10Hz)
Injection at 0.3x, 1x, 3x KD (100nM from equilibrium binding experiments)
*We suspect the analyte may be a non-covalent dimer*
1) sticking to flow cells? aggregated protein? (fresh off the SEC S200 column though!)
2) why is my control cell giving a higher signal in the dissociation phase? can I set this as zero to calculate the Kd?
3) two-phase dissociation (evidence of 2:1 stoichiometry, maybe?)
In short, is there useable data here?
Any help would be greatly appreciated!
Lee
BIAcore 3000, CM5 chip, amine-coupled streptavidin, biotinylated ligands.
FC1 7310 strep + 140 control protein (45kDa)
FC2 6753 strep + 302 ligand protein (75kDa)
100 ul/min, KINJECT 25ul, 90s dissociation
single flow cell injections at maximal data rate (10Hz)
Injection at 0.3x, 1x, 3x KD (100nM from equilibrium binding experiments)
*We suspect the analyte may be a non-covalent dimer*
1) sticking to flow cells? aggregated protein? (fresh off the SEC S200 column though!)
2) why is my control cell giving a higher signal in the dissociation phase? can I set this as zero to calculate the Kd?
3) two-phase dissociation (evidence of 2:1 stoichiometry, maybe?)
In short, is there useable data here?
Any help would be greatly appreciated!
Lee
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
10 years 8 months ago #2
by Arnoud
Replied by Arnoud on topic Odd kinetic trace
Hi Lee,
Let’s start with 2)
The higher control channels seems not to bad. I would try subtracting it.
I am more concerned about how the whole curve looks like: partly fast association and dissociation and partly slow dissociation. Clearly something in the system is not working.
The reference seems to compensate, but further optimization is needed.
1) sticking could be possible. Try some extra salt or detergent.
3) Based on these sensorgrams I would not think of that. It is better to use other techniques to determine the stoichiometry like ultracentrifugation or size exclusion chromatography
Arnoud
Let’s start with 2)
The higher control channels seems not to bad. I would try subtracting it.
I am more concerned about how the whole curve looks like: partly fast association and dissociation and partly slow dissociation. Clearly something in the system is not working.
The reference seems to compensate, but further optimization is needed.
1) sticking could be possible. Try some extra salt or detergent.
3) Based on these sensorgrams I would not think of that. It is better to use other techniques to determine the stoichiometry like ultracentrifugation or size exclusion chromatography
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud