This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
reproducibility of sensograms and general data quality
- sabyasachisen
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago #1
by sabyasachisen
reproducibility of sensograms and general data quality was created by sabyasachisen
hello,
for quite sometime i have been following the forum discussions, and as a beginner in SPR application, i have benefited a lot. i am studying protein-peptide interaction using BIACORE X100, with CM5 chip, immobilized with anti-GST Ab. the ligand (GST tagged protein) is captured on to this surface. due to lack of expertise, i am unable to troubleshoot some problems. :
1. firstly, my data is not reproducible. even within a particular kinetic experiment, the repeat sensograms do not overlap (i.e. sensograms of the repeated conc., i use a multicycle kinetics approach). i doubt if my regeneration is working fine. i use 2 successive injections of 10mM glycine -HCL + 30% ethylene glycol) with a contact time of 60 sec each.
2. secondly, i get huge injection spikes before and after the analyte injection. i have tried varying the injection rate. i highest i tried being 50ul/min, but the problem persists.
3. finally, it would be of great help, if i could get some comments on the overall quality of the sensograms, and any suggestion to improve the quality of assays.
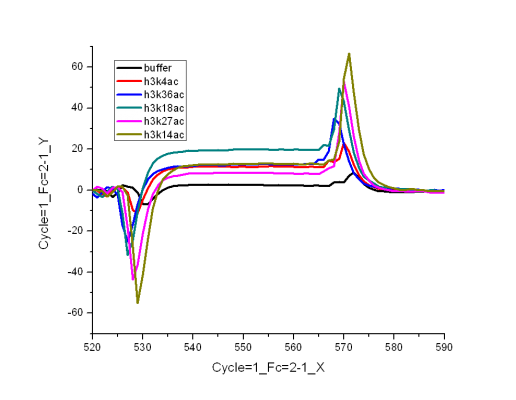
this represents the buffer spikes
...and this one shows the two same conc. of analytes not overlaying
also, i seem to have a problem of analyte dissociation during the injection phase itself, as it appears from the plot above. i was wondering what could be the reason for this.
for quite sometime i have been following the forum discussions, and as a beginner in SPR application, i have benefited a lot. i am studying protein-peptide interaction using BIACORE X100, with CM5 chip, immobilized with anti-GST Ab. the ligand (GST tagged protein) is captured on to this surface. due to lack of expertise, i am unable to troubleshoot some problems. :
1. firstly, my data is not reproducible. even within a particular kinetic experiment, the repeat sensograms do not overlap (i.e. sensograms of the repeated conc., i use a multicycle kinetics approach). i doubt if my regeneration is working fine. i use 2 successive injections of 10mM glycine -HCL + 30% ethylene glycol) with a contact time of 60 sec each.
2. secondly, i get huge injection spikes before and after the analyte injection. i have tried varying the injection rate. i highest i tried being 50ul/min, but the problem persists.
3. finally, it would be of great help, if i could get some comments on the overall quality of the sensograms, and any suggestion to improve the quality of assays.
this represents the buffer spikes
...and this one shows the two same conc. of analytes not overlaying
also, i seem to have a problem of analyte dissociation during the injection phase itself, as it appears from the plot above. i was wondering what could be the reason for this.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
11 years 9 months ago #2
by Arnoud
Replied by Arnoud on topic reproducibility of sensograms and general data quality
Hi,
A quick answer
1) Regeneration is different for every ligand and antigen pair. So you have to optimize the regeneration every time. You can start with reading www.sprpages.nl/kinetics/regeneration.html
Also I think a good regeneration has the shortest contact time possible.
2) Use the best inject command available and in your case with the regeneration with glycol use extra wash steppes!
3) Non reproducible duplo's can be cause by several things.
- regeneration is not optimal --> baseline wil be higher every cycle
- regeneration is detrimental to your ligand --> less analyte binding
- analyte is binding to the vial --> actual concentration is lower and thus the curve
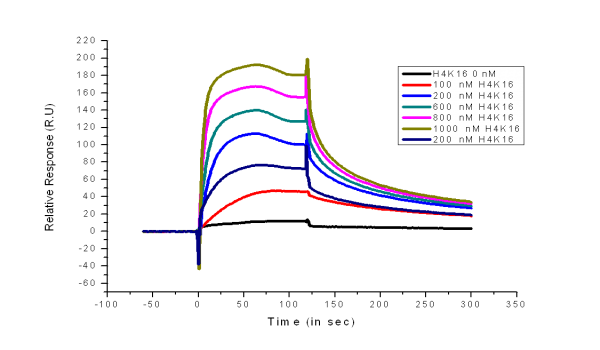
The 'dissociation' during injection is some times puzzeling. Some times it is the injection system which is not clean enough (buffer and sample are not well separated) and some times it seems that there is some rearrangement on the chip. As you notice it is concentration dependent.
The other odd thing is, that the curves are evenly spaced between 200 and 1000 nM of H4K16.
How is your blank doing in this setup?
I suggest you read the sensorgram tutorial at www.sprpages.nl/sensorgram-tutorial.html and make the quiz!
To have a guide to setup your experiments, download the Checklist Experiments from the Downloadspage: www.sprpages.nl/downloads/methods.html
Keep us informed about your progress.
Kind regards
Arnoud
A quick answer
1) Regeneration is different for every ligand and antigen pair. So you have to optimize the regeneration every time. You can start with reading www.sprpages.nl/kinetics/regeneration.html
Also I think a good regeneration has the shortest contact time possible.
2) Use the best inject command available and in your case with the regeneration with glycol use extra wash steppes!
3) Non reproducible duplo's can be cause by several things.
- regeneration is not optimal --> baseline wil be higher every cycle
- regeneration is detrimental to your ligand --> less analyte binding
- analyte is binding to the vial --> actual concentration is lower and thus the curve
The 'dissociation' during injection is some times puzzeling. Some times it is the injection system which is not clean enough (buffer and sample are not well separated) and some times it seems that there is some rearrangement on the chip. As you notice it is concentration dependent.
The other odd thing is, that the curves are evenly spaced between 200 and 1000 nM of H4K16.
How is your blank doing in this setup?
I suggest you read the sensorgram tutorial at www.sprpages.nl/sensorgram-tutorial.html and make the quiz!
To have a guide to setup your experiments, download the Checklist Experiments from the Downloadspage: www.sprpages.nl/downloads/methods.html
Keep us informed about your progress.
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud