Declining binding signal during injection
- Helge
- Topic Author
- New Member
-

- Thank you received: 0
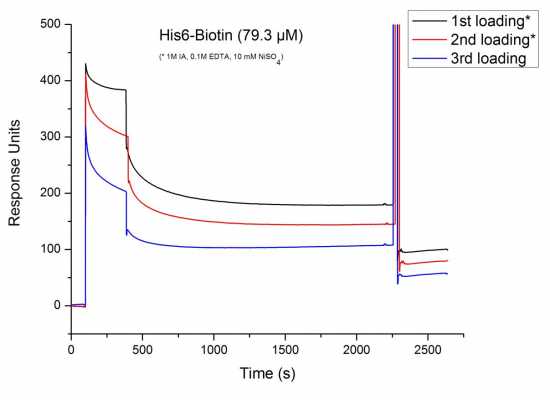
I did a very simple binding study in which a short hexahistidine-biotin peptide was injected onto an SPR chip functionalized with a bisNTA-SAM (I used bisNTA thiols like this: www.rsc.org/ej/CS/2011/c0cs00056f/c0cs00056f-f18.gif ).
Experimental conditions:
Here are the results:
The peptide clearly binds to the surface. There is quite some non-specific binding, but that is not what bothers me. My problem is that the peptide dissociates already during the injection phase (the dissociation seems to get stronger with each cycle, but this can be attributed to non-specifically bound peptides still remaining on the surface). Additionally, this effect seems to be concentration dependent, i.e. lower concentrations show a less pronounced dissociation; for concentrations of <1 µM I get normal looking association curves (data not shown).
Has anybody an explanation for this?
I checked the literature and several mechanisms have been suggested:
Nickel leaching: After binding to the Ni-NTA complexes the His6-peptides disociate again and "steal" the Ni2+ ions from the chelators, steadily reducing the number of binding sites during the injection phase.
I'm not entirely sure that this is what happened here. I did a lot of SPR with His6-Proteins on bisNTA-SAMs in the past and I never saw this kind of association curve or a general decline in binding signal after repeated injection cycles. On the other hand, I usally used concentration in the sub-micromolar range when working with proteins. Here I work with around 80 µM...
Rebinding effects:Analytes dissociate from a ligand but quickly bind to another one. These events do not show up in the binding curve and give the impression of a binding affinity that seems stronger than it actually is. For higher concentrations, however, there are less free binding sites for a dissociated analyte to rebind to causing a stronger decline of the binding curve during dissociation.
Not sure of this being the reason for my results, either. Rebinding can explain differences in the dissociation kinetics for different analyte concentrations. But I have a hard time understanding how rebinding effects could lead to a decrease in binding signal during association. As long as there are still free binding sites the signal should keep increasing. Once the maximum number of binding sites is occupied (i.e. equilibrium is reached) the signal should be stable, regardless of any rebinding. On top of that, this experiment was done on a chip functionalized with a SAM, i.e. a 2D system. Once a protein dissociates it will move away from the surface. Unlike
on dextrane matrix substrates, which are 3D systems, proteins will rarely rebind on SAMs, I think.
Here is another idea: BisNTA contains two binding sites for His-Tags. Since it can bind the same His-Tag with two NTAs at the same time this interaction is much stronger than the monoNTA-HisTag interaction. However, let's say we inject a high concentration of His-peptides. We can assume that shortly after the injection the surface is saturated with bound peptides. Let's assume further that some bisNTAs have bound two His-peptides, one peptide on each NTA, that now both compete with each other for a bi-valent binding. One peptide will eventually win and and strongly bind to both NTAs, thereby displacing the other peptide. In the early injection phase this process will happen more often, while in the late phase only a few bisNTAs with two transiently bound peptides are left. This would explain the exponential-like decline of the binding signal during injection.
As for the concentration dependence: For lower peptide concentrations this effect will be weaker because it will be less probable for a bisNTA to bind two peptides at the same time. If a peptide comes along and binds to one of the two NTAs it will quickly bind to the other one as well because there is no other peptide around that could interfere. Therefore the binding curves look "normal" for lower concentrations.
Does this make sense? Are there other mechanisms that could cause such curves that I am missing here? What do you guys think?
Thanks for your input!

Helge
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

- Thank you received: 0
I don't think that there is a competion between the two binding sites on one BisNTA. The binding site are equal in strength. You can compare them with an IgG antibody. Since you not state the size of the his-peptide-biotin it is not possible to know if one peptide is blocking more than one NTA-site.
Do you know how pure your peptide is? If there is a population of smaller peptides it is possible that these are competing for binding.
Looking at your sensorgram I think you have a decent response. Regeneration can be better..
What is your goal with the biotin?
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- Helge
- Topic Author
- New Member
-

- Thank you received: 0
thanks for your reply.
Hm, could you elaborate on that? Rebinding would make perfect sense for me if I would see concentration dependent differences during dissociation. But how can rebinding lead to dissociation during the injection phase?I think is has more to do with the rebinding effects. I agree with your paragraph.
Just to make sure we're not talking past each other: What I was trying to say was that two losely bound peptides (one His-tag on each NTA) compete with each other for being bound to both NTAs (one His-tag on both NTAs, the other one displaced). Because of "cooperativity effects" the bivalent interaction is much stronger than the sum of two monovalent interactions. So in a situation in which two peptides are losely bound to the bisNTA each peptide wants the whole bisNTA for itself. At some point one will win over the other, kicking it out. And since the binding is now much stronger other peptides in the solution will not be able to displace the winner easily. Please tell me if I made a mistake in my reasoning.I don't think that there is a competion between the two binding sites on one BisNTA. The binding site are equal in strength. You can compare them with an IgG antibody.
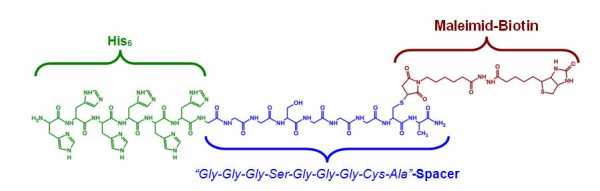
You're right. I should have been more specific here. The peptide is pretty small. Please see the picture below. If the hexahistidine is stably bound to the bisNTA it will definitely occupy both NTAs.Since you not state the size of the his-peptide-biotin it is not possible to know if one peptide is blocking more than one NTA-site.
It should be very pure. HPLC chromatograms and mass spec data looked fine.Do you know how pure your peptide is? If there is a population of smaller peptides it is possible that these are competing for binding.
Yes, regeneration is a bit disappointing. I blame the biotin for that.Looking at your sensorgram I think you have a decent response. Regeneration can be better.
I wanted to immobilize Neutravidin-conjugated FluoSpheres on bisNTA-functionalized gold surfaces. After activating the bisNTA surface with the His-Biotin peptide I wanted to add the FluoSpheres. The SPR experiment should tell me how much piptide can be bound on the surface and how stable it binds. But when I saw how bad regeneration was I instead chose to activate the NA-FluoSpheres with the peptide and immobilize these complexes on the bisNTA surface. In the end this worked out much better.What is your goal with the biotin?
Anyway, during the SPR run I saw these strange curves. It's puzzling me and I'm looking for a reasonable explanation.
Thanks so much,
Helge
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

- Thank you received: 0
I think is has more to do with the rebinding effects. I agree with your paragraph.
Hm, could you elaborate on that? Rebinding would make perfect sense for me if I would see concentration dependent differences during dissociation. But how can rebinding lead to dissociation during the injection phase?
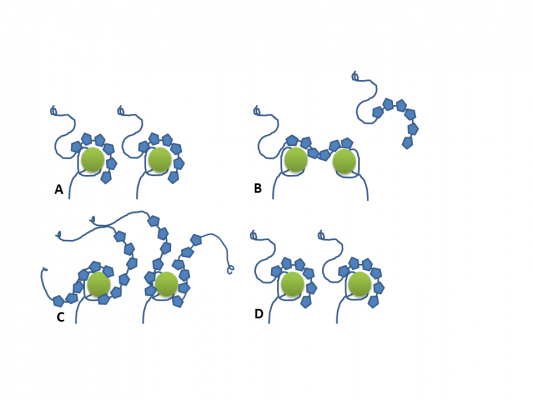
Ok, thinking about it, it is probably not rebinding in the way we use it normally. If you look at the pictures below I would call it rearranging. Due to the high concentration of the 6xHIS-peptide, initially there are more than one 6x-HIS peptide bound to one NiNTA-moeity. When one of the peptides binds stronger it will displace the other, lowering the amount of bound peptide. This looks consistent with the observation that this is not so pronounced at lower peptide concentrations.
In a cartoon.
A --> B is what you propose. What you say sounds plausible, but I am not aware of any proof that the 6xHIS is capable to bind to two NiNTA-moieties. I think the cartoons C --> D are more plausible. However, it is difficult to proof!
Looking at your sensorgram I think you have a decent response. Regeneration can be better.
Yes, regeneration is a bit disappointing. I blame the biotin for that.
You can use 0.35M EDTA to strip the Nickel from the NTA. This will strip all positive charges from the surface. The drawback is that you have to add new Nickel to 'activate' the NTA.
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- Helge
- Topic Author
- New Member
-

- Thank you received: 0
Good point, I'm not aware of a proof for this, either. However, the two NTAs are very close so sterically it should be absolutely possible for both NTAs to bind the same His6. Lata et al. synthesized a trisNTA and a tetrakisNTA with much higher affinities than bisNTA and they explain this by simultaneous binding of all NTA-groups to the same oligo-His peptide (J. Am. Chem. Soc. 127 (2005) 10205-15).A --> B is what you propose. What you say sounds plausible, but I am not aware of any proof that the 6xHIS is capable to bind to two NiNTA-moieties.
Well, actually it might be entirely possible that both case are happening.I think the cartoons C --> D are more plausible. However, it is difficult to proof!
Looking at your sensorgram I think you have a decent response. Regeneration can be better.
Yes, regeneration is a bit disappointing. I blame the biotin for that.
Yes, we did that as well (although we used 0.1M instead) but it didn't work either. I guess the peptide just didn't like the surface.You can use 0.35M EDTA to strip the Nickel from the NTA. This will strip all positive charges from the surface. The drawback is that you have to add new Nickel to 'activate' the NTA.
Again, thanks for your input on this.
Cheers,
Helge
Please Log in or Create an account to join the conversation.