This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
Protein immobilization
- Jenny
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 2 months ago - 11 years 2 months ago #1
by Jenny
Protein immobilization was created by Jenny
Hi,
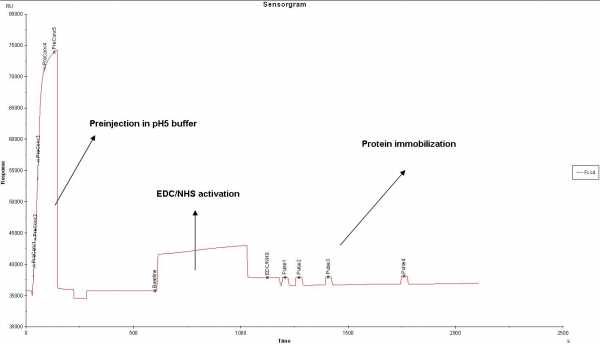
Recently we have a problem with our protein study. We have a protein 5mg/ml in 40mM Bistris pH 7.8; 10% Glycerol, 200mM NaCl, 0.5mM DTT (For SPR assay, we only use 10 ug/ml protein). We already did the pH scouting and found pH5 acetate buffer is the best. However, when we use general CM5 amine coupling procedure, we found that the protein cannot be immobilized onto the chip. The strange thing is when we do preinjection, the protein could be preconcentrated onto the chip ~35000 RU. After EDC/NHS activation, when we inject the protein again, the curve went almost flat, no preconcentration nor immobilization (I have attached the curve blow). Could anyone give some suggestion? Thanks a lot for your help!
Recently we have a problem with our protein study. We have a protein 5mg/ml in 40mM Bistris pH 7.8; 10% Glycerol, 200mM NaCl, 0.5mM DTT (For SPR assay, we only use 10 ug/ml protein). We already did the pH scouting and found pH5 acetate buffer is the best. However, when we use general CM5 amine coupling procedure, we found that the protein cannot be immobilized onto the chip. The strange thing is when we do preinjection, the protein could be preconcentrated onto the chip ~35000 RU. After EDC/NHS activation, when we inject the protein again, the curve went almost flat, no preconcentration nor immobilization (I have attached the curve blow). Could anyone give some suggestion? Thanks a lot for your help!
Last edit: 11 years 2 months ago by Jenny. Reason: need to add more conditions
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
11 years 2 months ago #2
by Arnoud
Replied by Arnoud on topic Protein immobilization
Hi Jenny,
It is puzzeling what happend. First, I think the pre-concentration with 10 ug/ml ligand is very high. It can be an effect of the glycerol (0.02%). Are you aiming for a high density surface?
How was the pH scouting? Was it high for more solutions?
Also, how big is your ligand?
As an remark: a high pre-concentration is no garantee for a high immobilization (covalent-bond).
It is puzzeling what happend. First, I think the pre-concentration with 10 ug/ml ligand is very high. It can be an effect of the glycerol (0.02%). Are you aiming for a high density surface?
How was the pH scouting? Was it high for more solutions?
Also, how big is your ligand?
As an remark: a high pre-concentration is no garantee for a high immobilization (covalent-bond).
Please Log in or Create an account to join the conversation.
- Jenny
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 2 months ago #3
by Jenny
Replied by Jenny on topic Protein immobilization
Hi Arnoud,
Thanks a lot for your reply! Yes, we want a high density surface to test small analytes (MW~400). The ligand is ~40K. For pH scouting we tested 4.5,5.0, 5.5, and 5.0 gave the highest level. The problem is why after NHS/EDC activation, the protein cannot accumulate onto the surface.
Jenny
Thanks a lot for your reply! Yes, we want a high density surface to test small analytes (MW~400). The ligand is ~40K. For pH scouting we tested 4.5,5.0, 5.5, and 5.0 gave the highest level. The problem is why after NHS/EDC activation, the protein cannot accumulate onto the surface.
Jenny
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
11 years 2 months ago #4
by Arnoud
Replied by Arnoud on topic Protein immobilization
Hi Jenny,
Some thoughts:
- Did you do the immobilization by hand or did you let the wizard do the immobilization? If using the wizard, It is possible that it will not work due to the high preconcentration rate. Try immobilization by hand to control pre-concentration and monitor coupling.
- Is it possible do get rid (dialyse out) of the glycerol and DTT before immobilization? Immobilization could be better without.
- Longer activation times wil activate the sensor chip surface more. Since you need a lot of ligand on the chip, this can be helpful.
- check your chemicals and buffer (make fresh)
Arnoud
Some thoughts:
- Did you do the immobilization by hand or did you let the wizard do the immobilization? If using the wizard, It is possible that it will not work due to the high preconcentration rate. Try immobilization by hand to control pre-concentration and monitor coupling.
- Is it possible do get rid (dialyse out) of the glycerol and DTT before immobilization? Immobilization could be better without.
- Longer activation times wil activate the sensor chip surface more. Since you need a lot of ligand on the chip, this can be helpful.
- check your chemicals and buffer (make fresh)
Arnoud
Please Log in or Create an account to join the conversation.
- Helge
- New Member
-

Less
More
- Thank you received: 0
11 years 1 month ago - 11 years 1 month ago #5
by Helge
In addition to that, you might want to test another buffer system. BisTris contains a lot of hydroxyl groups which have been observed to react well with NHS ester, too. ( chemistry.stackexchange.com/questions/42...ons-of-nhs-chemistry ).
Try Hepes or PBS buffers.
Good luck!
Replied by Helge on topic Protein immobilization
Arnoud wrote: - check your chemicals and buffer (make fresh)
In addition to that, you might want to test another buffer system. BisTris contains a lot of hydroxyl groups which have been observed to react well with NHS ester, too. ( chemistry.stackexchange.com/questions/42...ons-of-nhs-chemistry ).
Try Hepes or PBS buffers.
Good luck!

Last edit: 11 years 1 month ago by Helge. Reason: Typo
Please Log in or Create an account to join the conversation.
- Jenny
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 1 month ago #6
by Jenny
Replied by Jenny on topic Protein immobilization
Hi, Arnoud and Helge,
Thanks a lot!
I will try manual injection to see whether I can get better result.
Best,
Jenny
Thanks a lot!
I will try manual injection to see whether I can get better result.
Best,
Jenny
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud