These are the posts of the old forum. It was not possible to transfer the user data, so they are missing in most of the posts. For new questions, go to the general discussions.
Non-specific binding
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #1
by OldForum
Non-specific binding was created by OldForum
I am unable to get a good Langmuir fitting and I wonder if this is because of the non-specific binding in the reference cell being different from any non-specific binding that I might have in my ligand-coupled cell.
I see a non-specific binding of about 130RU at highest concentration of my analyte (1 uM) to the activated-ethanolamine blocked reference cell.
This non-specific binding leaves behind about 70 RU signal after 300 second of dissociation.
Can I go ahead to use this signal as a reference cell or should I try ways to reduce the non-specific binding.
What can I do to further reduce this non-specific binding.
Can this non-specific binding contribute to a poor fitting.
patnml
I see a non-specific binding of about 130RU at highest concentration of my analyte (1 uM) to the activated-ethanolamine blocked reference cell.
This non-specific binding leaves behind about 70 RU signal after 300 second of dissociation.
Can I go ahead to use this signal as a reference cell or should I try ways to reduce the non-specific binding.
What can I do to further reduce this non-specific binding.
Can this non-specific binding contribute to a poor fitting.
patnml
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #2
by OldForum
Replied by OldForum on topic Non-specific binding
Strategies to reduce non-specific binding:
-- add more P20, e.g. up to 0.1 %
-- add more NaCl, e.g. up to 0.5 M
-- add CM dextran, e.g. up to 0.1 mg/ml
-- immobilize a non related protein to the reference channel. BSA is often used but can give more non-specific binding.
Because your reference cell does not contain protein non-specific binding can be different from your positive channels.
You can try buffer injections to determine if there is a difference between the channels. If buffer injections already give you differences you should try to do a double referencing, meaning that you subtract blank injection from the signal after you subtracted the reference cell to compensate for channel behaviour.
It is hard to tell what you mean with a poor fitting without seeing the curves. In general, low non-specific binding will give better curves to resolve but the there are so much other things that can influence the binding that it is hard to say that is only non-specific binding dependent.
I hope this will help you a little further
Arnoud
-- add more P20, e.g. up to 0.1 %
-- add more NaCl, e.g. up to 0.5 M
-- add CM dextran, e.g. up to 0.1 mg/ml
-- immobilize a non related protein to the reference channel. BSA is often used but can give more non-specific binding.
Because your reference cell does not contain protein non-specific binding can be different from your positive channels.
You can try buffer injections to determine if there is a difference between the channels. If buffer injections already give you differences you should try to do a double referencing, meaning that you subtract blank injection from the signal after you subtracted the reference cell to compensate for channel behaviour.
It is hard to tell what you mean with a poor fitting without seeing the curves. In general, low non-specific binding will give better curves to resolve but the there are so much other things that can influence the binding that it is hard to say that is only non-specific binding dependent.
I hope this will help you a little further
Arnoud
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago - 11 years 3 months ago #3
by OldForum
Replied by OldForum on topic Non-specific binding
Hi Arnoud,
The suggestions have been most useful. I changed my buffer to increase NaCl (from 50 to 150 mM) and to add Tween-20 (0.05%).
This reduced NSB in the activated-ethanolamine-blocked channel to half without loss of binding in the ligand-immobilized channel (in fact there was an increase in binding with the new buffer in the ligand-immobilized channel).
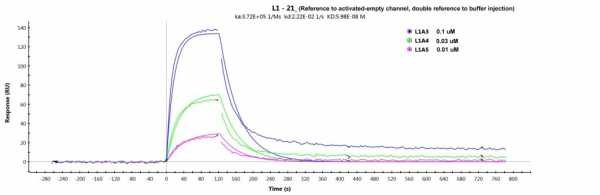
But when I try to do a Langmuir fitting with this new buffer and double referenced to the activated-ethanolamine-blocked channel, I still get a bad fitting (chi2=77, see fitting below).
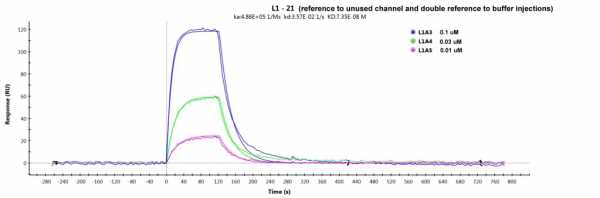
Surprising, changing the 1st referencing to an unused channel, gave a better fit (chi2 of 7, see fitting below)
Since the ligand-immobilized channel is also activated, shouldn't an activated-empty channel provide a better referencing? Why is the fit much worse? Is the better fit in the second result just a random event? Should I trust it?
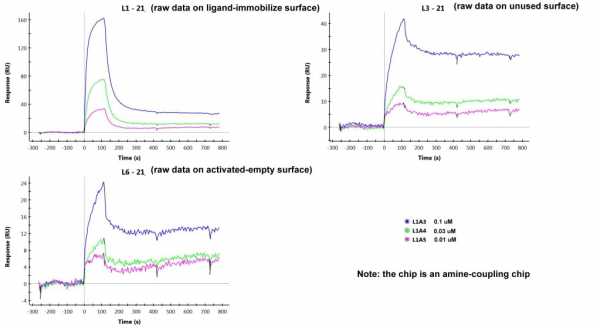
The difference between the unused channel and the activated-empty channel is that the NSB in an unused channel is higher than in the activated-empty channel (see raw data below).
patnml
The suggestions have been most useful. I changed my buffer to increase NaCl (from 50 to 150 mM) and to add Tween-20 (0.05%).
This reduced NSB in the activated-ethanolamine-blocked channel to half without loss of binding in the ligand-immobilized channel (in fact there was an increase in binding with the new buffer in the ligand-immobilized channel).
But when I try to do a Langmuir fitting with this new buffer and double referenced to the activated-ethanolamine-blocked channel, I still get a bad fitting (chi2=77, see fitting below).
Surprising, changing the 1st referencing to an unused channel, gave a better fit (chi2 of 7, see fitting below)
Since the ligand-immobilized channel is also activated, shouldn't an activated-empty channel provide a better referencing? Why is the fit much worse? Is the better fit in the second result just a random event? Should I trust it?
The difference between the unused channel and the activated-empty channel is that the NSB in an unused channel is higher than in the activated-empty channel (see raw data below).
patnml
Last edit: 11 years 3 months ago by OldForum.
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #4
by OldForum
Replied by OldForum on topic Non-specific binding
Hi Patnml,
Sorry for the delay but I was away.
Great that you have better result now! Since adding salt and Tween did make the curves better indicates that the non-specific binding comes from some ionic interaction. You can try to add more salts in steps of 10 mM to look at the effect on the curves.
The reason to activate and deactivate a reference channel is because the -COOH group has a higher charge than the -OH group of the ethanolamine. And that is what you see in the last figure, it is lower. Maybe increasing the activation time will activate more and deactivate more lowering the non-specific binding further. You can als use a sensor chip CM4 with a low carboxylation.
What do you mean with an activated-empty channel? An activated channel must be deactivated with ethanol amine or another substance which is reactive (e.g. TRIS). Because otherwise the activated channel will bind analyte wich will be covalent bound to the surface.
If you are interested in the K you can try the steady state affinity method. It wil not give you the full kinetics but can be helpfull in comparing batches.
Arnoud
Sorry for the delay but I was away.
Great that you have better result now! Since adding salt and Tween did make the curves better indicates that the non-specific binding comes from some ionic interaction. You can try to add more salts in steps of 10 mM to look at the effect on the curves.
The reason to activate and deactivate a reference channel is because the -COOH group has a higher charge than the -OH group of the ethanolamine. And that is what you see in the last figure, it is lower. Maybe increasing the activation time will activate more and deactivate more lowering the non-specific binding further. You can als use a sensor chip CM4 with a low carboxylation.
What do you mean with an activated-empty channel? An activated channel must be deactivated with ethanol amine or another substance which is reactive (e.g. TRIS). Because otherwise the activated channel will bind analyte wich will be covalent bound to the surface.
If you are interested in the K you can try the steady state affinity method. It wil not give you the full kinetics but can be helpfull in comparing batches.
Arnoud
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
13 years 8 months ago #5
by OldForum
Replied by OldForum on topic Non-specific binding
Hi Arnoud,
Good to have you back.
In my earlier post, activated-empty channel meant channels that are "activated by NHS:EDC and then blocked by ethanolamine".
Thank you for the explanation on COOH versus OH on the chip surface and for your suggestion to activate and deactivate more.
I will try that in my next experiment.
Hopefully I will bring better results to the forum the next time.
patnml
Good to have you back.
In my earlier post, activated-empty channel meant channels that are "activated by NHS:EDC and then blocked by ethanolamine".
Thank you for the explanation on COOH versus OH on the chip surface and for your suggestion to activate and deactivate more.
I will try that in my next experiment.
Hopefully I will bring better results to the forum the next time.
patnml
Please Log in or Create an account to join the conversation.