These are the posts of the old forum. It was not possible to transfer the user data, so they are missing in most of the posts. For new questions, go to the general discussions.
Problems with immobilisation
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago #1
by OldForum
Problems with immobilisation was created by OldForum
Hello,
As someone still learning SPR, I am having a tremendous amount of difficulty getting up and running. What follows is how I have completed my last two runs. The protocol I have been using is borrowed from the person who uses the machine, but they couple Neutravidin instead.
-Dock new chip (CM5)
-Prime
-Normalize with 40% Glycerol
-Run sensorgram (flow at 10ul/min)
- Inject 1:1 (0.10M NHS) (0.40M EDC) using kinject
100ul, 60s dissociation time
- Inject protein of interest to couple (have tried 30-50ug/ml)
-100ul
-60s dissociation time
About the protein: 16kD PI = 8.13 6x His tag, strep II tag 8 Lysines, 0 Cysteines
When scouting for immobilization pH, I have tried everything from Sodium Acetate pH 5.2 to 6.0, and ultrapure H20. There is never any difficulty achieving an RU difference of +12,000-24,000. Upon activating the surface however, the curve essentially flatlines and will barely achieve an RU difference of 3500, which will decrease rather steadily throughout the rest of the run.
Sometime I continue on with blocking, but since it appears that my protein is not actually being coupled, I usually end the run in frustration. If anyone has any useful suggestions or critiques, I would love to hear them. I have tried using Ni-NTA chips with a different system and had similar results. At this point, however, I am open to anything.
Thank you!
Claude Bessy
As someone still learning SPR, I am having a tremendous amount of difficulty getting up and running. What follows is how I have completed my last two runs. The protocol I have been using is borrowed from the person who uses the machine, but they couple Neutravidin instead.
-Dock new chip (CM5)
-Prime
-Normalize with 40% Glycerol
-Run sensorgram (flow at 10ul/min)
- Inject 1:1 (0.10M NHS) (0.40M EDC) using kinject
100ul, 60s dissociation time
- Inject protein of interest to couple (have tried 30-50ug/ml)
-100ul
-60s dissociation time
About the protein: 16kD PI = 8.13 6x His tag, strep II tag 8 Lysines, 0 Cysteines
When scouting for immobilization pH, I have tried everything from Sodium Acetate pH 5.2 to 6.0, and ultrapure H20. There is never any difficulty achieving an RU difference of +12,000-24,000. Upon activating the surface however, the curve essentially flatlines and will barely achieve an RU difference of 3500, which will decrease rather steadily throughout the rest of the run.
Sometime I continue on with blocking, but since it appears that my protein is not actually being coupled, I usually end the run in frustration. If anyone has any useful suggestions or critiques, I would love to hear them. I have tried using Ni-NTA chips with a different system and had similar results. At this point, however, I am open to anything.
Thank you!
Claude Bessy
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago - 10 years 2 weeks ago #2
by OldForum
Replied by OldForum on topic Problems with immobilisation
Hi Claude,
I have some remarks and questions for you.
Pre-concentration is meant to stimulate a high local ligand concentration at the sensor chip surface. A high pre-concentration is no guarantee for a high immobilization. Even a fast pre-concentration will not help you. I use the following protocol.
flow: 5 ul/min
Activation: quickinject with 35 ul NHS/EDC 1:1 (contact time 7 minutes).
Ligand: 10 ug/ml in 10 mM Actetate buffer pH 5.5 (pH depending on protein)
Use manual inject to adjust the amount of ligand bound.
Deactivation: quickinject with 35 ul 1 M ethanolamine pH 8.0 (or use a diluted solution or TBS).
If your ligand is 16 kDa, how big is your analyte?
What is the goal of your experiment? Read www.sprpages.nl/immobilization.html
Can you post a sensorgram of your immobilization?
You do not need a high analyte response to do kinetics. Read www.sprpages.nl/sensorgram-tutorial.html
A difference of 3500 RU is high enough if you use a bigger analyte. Even an analyte of 250 Da should be measurable with this amount of ligand (assuming 100% is active).
You can use the BiaCalculations to calculate the amount of ligand you should immobilize for a proper response. Look at www.sprpages.nl/downloads/software.html
kind regards
Arnoud
I have some remarks and questions for you.
Pre-concentration is meant to stimulate a high local ligand concentration at the sensor chip surface. A high pre-concentration is no guarantee for a high immobilization. Even a fast pre-concentration will not help you. I use the following protocol.
flow: 5 ul/min
Activation: quickinject with 35 ul NHS/EDC 1:1 (contact time 7 minutes).
Ligand: 10 ug/ml in 10 mM Actetate buffer pH 5.5 (pH depending on protein)
Use manual inject to adjust the amount of ligand bound.
Deactivation: quickinject with 35 ul 1 M ethanolamine pH 8.0 (or use a diluted solution or TBS).
If your ligand is 16 kDa, how big is your analyte?
What is the goal of your experiment? Read www.sprpages.nl/immobilization.html
Can you post a sensorgram of your immobilization?
You do not need a high analyte response to do kinetics. Read www.sprpages.nl/sensorgram-tutorial.html
A difference of 3500 RU is high enough if you use a bigger analyte. Even an analyte of 250 Da should be measurable with this amount of ligand (assuming 100% is active).
You can use the BiaCalculations to calculate the amount of ligand you should immobilize for a proper response. Look at www.sprpages.nl/downloads/software.html
kind regards
Arnoud
Last edit: 10 years 2 weeks ago by .
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago #3
by OldForum
Replied by OldForum on topic Problems with immobilisation
Arnuod,
As always, thank you for the reply.
The analyte I will be studying is a 7AA peptide. I will also be studying larger peptides, but this will serve as my "baseline" for measuring affinity to my protein.
Our lab paid a company to do the prior protein-peptide SPR experiment and I thought (foolishly) that I would try to emulate their protocol. They were able to bind to 6500RU.
I will post a sensorgram when I get to the lab.
One of the strange things I noticed the other day is that when I blocked with Ethanolamine (the RU indicated a signal above the baseline after activation) was that the signal did not increase at all. Even in theory if all the activated groups bound protein, wouldn't there still be a signal increase?
I will try using quickinject for my next run. Like I mentioned before, I have been mainly using a protocol that has worked for someone doing completely different work on the machine. But I feel like I am learning gradually. Thanks again for the reply.
As always, thank you for the reply.
The analyte I will be studying is a 7AA peptide. I will also be studying larger peptides, but this will serve as my "baseline" for measuring affinity to my protein.
Our lab paid a company to do the prior protein-peptide SPR experiment and I thought (foolishly) that I would try to emulate their protocol. They were able to bind to 6500RU.
I will post a sensorgram when I get to the lab.
One of the strange things I noticed the other day is that when I blocked with Ethanolamine (the RU indicated a signal above the baseline after activation) was that the signal did not increase at all. Even in theory if all the activated groups bound protein, wouldn't there still be a signal increase?
I will try using quickinject for my next run. Like I mentioned before, I have been mainly using a protocol that has worked for someone doing completely different work on the machine. But I feel like I am learning gradually. Thanks again for the reply.
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago #4
by OldForum
Replied by OldForum on topic Problems with immobilisation
I was wrong. I guess the last two coupling sensorgrams were not saved. I am scheduled for another run next Thursday, so maybe I will only use two of the flow cells on the chip and then see what happens.
Claude Bessy
Claude Bessy
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago #5
by OldForum
Replied by OldForum on topic Problems with immobilisation
Hi Claude,
The reason why I use the quickinject, is that it goes quicker and it makes no difference in the sensorgrams.
Use the manual inject to target the proper response. To get more on the sensor chip you can activate a litle longer (10 minutes).
A marginal response increase due to attachment of ethanolamine can be expected although a decrease in signal due to washing out of non covalent bound ligand (electrostatically bound) is also possible.
In general I use the immobilization response after deactivation as a guideline. The real amount of active ligand is hard to determine.
As you found out, recording and saving all of your data is important. You can look at www.sprpages.nl/downloads/methods.html to find out what is available.
kind regards
Arnoud
The reason why I use the quickinject, is that it goes quicker and it makes no difference in the sensorgrams.
Use the manual inject to target the proper response. To get more on the sensor chip you can activate a litle longer (10 minutes).
A marginal response increase due to attachment of ethanolamine can be expected although a decrease in signal due to washing out of non covalent bound ligand (electrostatically bound) is also possible.
In general I use the immobilization response after deactivation as a guideline. The real amount of active ligand is hard to determine.
As you found out, recording and saving all of your data is important. You can look at www.sprpages.nl/downloads/methods.html to find out what is available.
kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
11 years 9 months ago #6
by OldForum
Replied by OldForum on topic Problems with immobilisation
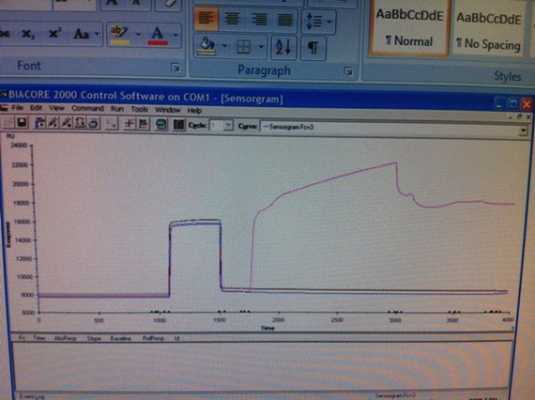
I attempted another run today. I injected:
35ul of EDC/NHS at 5ul/min using QUICKINJECT
100ul of ligand using QUICKINJECT
35ul Ethanolamine using QUICKINJECT
I am not sure about the efficiency of the ethanolamine deactivating the active groups. I am also not sure about the integrity of my protein at this point. I was able to couple a lot more this time, but I am not sure if this data is even correct.
Here is a copy of the sensorgram (sorry about the quality, I was not able to pull data off the machine itself).
Thanks for any suggestions anyone can provide. More information can also be provided.
35ul of EDC/NHS at 5ul/min using QUICKINJECT
100ul of ligand using QUICKINJECT
35ul Ethanolamine using QUICKINJECT
I am not sure about the efficiency of the ethanolamine deactivating the active groups. I am also not sure about the integrity of my protein at this point. I was able to couple a lot more this time, but I am not sure if this data is even correct.
Here is a copy of the sensorgram (sorry about the quality, I was not able to pull data off the machine itself).
Thanks for any suggestions anyone can provide. More information can also be provided.
Please Log in or Create an account to join the conversation.