These are the posts of the old forum. It was not possible to transfer the user data, so they are missing in most of the posts. For new questions, go to the general discussions.
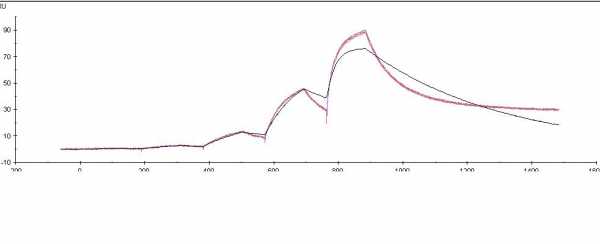
Biphasic curve
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 5 months ago #1
by OldForum
Biphasic curve was created by OldForum
Please Log in or Create an account to join the conversation.
- OldForum
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
12 years 5 months ago #2
by OldForum
Replied by OldForum on topic Biphasic curve
Biphasic means that there are more than one set of interactions going on. So the curves give you the total amount of response of interaction 1, interaction 2, interaction ....
The interaction can come from ligand and analyte heterogeneity (i.e. contamination with other proteins).
The interactions differ in association and dissociation rate. If you try to fit this with a 1:1 model it does not fit.
Fitting with a more complex model will make the fitting better, but do you know how to interpret the results? Which of the variables is your interaction? It is better to get rid of the heterogeneity.
Sometimes the interaction is with the dextran matrix but I assume you have corrected for that.
What you can try is to raise the NaCl concentration (e.g. 250 mM) to suppress low affinity interactions.
You can check the purity of the analyte on a gel and take action if not >95% pure.
If possible use a capture method to capture the ligand so it is uniformly orientated for interaction.
Sometimes it is difficult to solve this type of interaction. It will take time and effort to identify the source of the heterogeneity and to get rid of it.
kind regards
Arnoud
The interaction can come from ligand and analyte heterogeneity (i.e. contamination with other proteins).
The interactions differ in association and dissociation rate. If you try to fit this with a 1:1 model it does not fit.
Fitting with a more complex model will make the fitting better, but do you know how to interpret the results? Which of the variables is your interaction? It is better to get rid of the heterogeneity.
Sometimes the interaction is with the dextran matrix but I assume you have corrected for that.
What you can try is to raise the NaCl concentration (e.g. 250 mM) to suppress low affinity interactions.
You can check the purity of the analyte on a gel and take action if not >95% pure.
If possible use a capture method to capture the ligand so it is uniformly orientated for interaction.
Sometimes it is difficult to solve this type of interaction. It will take time and effort to identify the source of the heterogeneity and to get rid of it.
kind regards
Arnoud
Please Log in or Create an account to join the conversation.