This forum is intended for questions about kinetics, Surface Plasmon Resonance and the instruments related to these techniques.
can't perform calibration
- siddhartha2020
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
3 years 3 months ago #1
by siddhartha2020
can't perform calibration was created by siddhartha2020
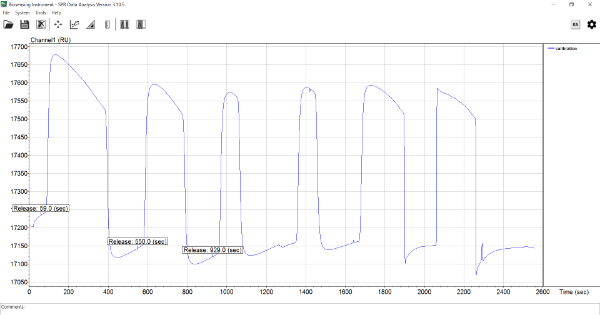
im using BI4500 SPR machine. but i tried to perform calibration several time. we also sent machine back. but company said macine is okay. but when i tried to calibrate , it gave unstable base line and resonse was decreasing. we tried to use this machine for Aptamer and protein binding. but we couldn't generate one results using BI4500. do anyone use this machine for aptamer and protein binding and also do you ever experience this kind of unstable baseline. if that could you please help me with this.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
3 years 3 months ago #2
by Arnoud
Replied by Arnoud on topic can't perform calibration
What do you mean with calibration? In general, SPR responses are differences between baseline and binding. I can see that the baseline is varying after the injections shown but what are you injecting? In addition, some internet searching reveals that the aptamer problem keeps popping up. Maybe it is time to start again and validate every step you take.
Validating the instrument.
-perform all the cleaning steps recommended by the manufacturer.
-make new fresh buffers etc. Filter and degas.
-dock new sensor chip (carboxy methylated dextran) and flow the buffer until stable base line (overnight).
-checking the injection system / detector by injecting an elevated salt solution (e.g. buffer + 25 mM added NaCl).
---this will show a jump in the sensor gram which must be square of shape.
-You can do a simple antibody – protein test on the sensor chip to show you can measure something
---this will test immobilization chemicals in case of an amine immobilization
---perform an ELISA when you suspect that immobilization is destroying the ligand
with kind regards
Arnoud
Validating the instrument.
-perform all the cleaning steps recommended by the manufacturer.
-make new fresh buffers etc. Filter and degas.
-dock new sensor chip (carboxy methylated dextran) and flow the buffer until stable base line (overnight).
-checking the injection system / detector by injecting an elevated salt solution (e.g. buffer + 25 mM added NaCl).
---this will show a jump in the sensor gram which must be square of shape.
-You can do a simple antibody – protein test on the sensor chip to show you can measure something
---this will test immobilization chemicals in case of an amine immobilization
---perform an ELISA when you suspect that immobilization is destroying the ligand
with kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- siddhartha2020
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
3 years 2 months ago #3
by siddhartha2020
Replied by siddhartha2020 on topic can't perform calibration
Hi Arnoud,
Thank you for your response.
i didn't understand what you told me that immobilization is destroying the ligand, could you please elaborate that to me. and could immobilization can destroy the ligand. how we use the ELISA test to check that.
Thank you for your response.
i didn't understand what you told me that immobilization is destroying the ligand, could you please elaborate that to me. and could immobilization can destroy the ligand. how we use the ELISA test to check that.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
3 years 2 months ago #4
by Arnoud
Replied by Arnoud on topic can't perform calibration
Hi,
I am not too familiar with aptamers but have you a way to detect them other than SPR? Maybe they are biotinylated on one side? Then coat the protein and see if you can detect aptamer binding (protein coat - aptamer-biotin - streptavidin-HRP detection).
The amine coupling makes covalent bonds between the ligand and sensor surface. When these bonds are at a spot used for binding the binding is not possible anymore.
In my last post there some questions which need answering to better understand what you tried. In addition, it is not clear if you immobilize the protein or the aptamer.
Kind regards
Arnoud
I am not too familiar with aptamers but have you a way to detect them other than SPR? Maybe they are biotinylated on one side? Then coat the protein and see if you can detect aptamer binding (protein coat - aptamer-biotin - streptavidin-HRP detection).
The amine coupling makes covalent bonds between the ligand and sensor surface. When these bonds are at a spot used for binding the binding is not possible anymore.
In my last post there some questions which need answering to better understand what you tried. In addition, it is not clear if you immobilize the protein or the aptamer.
Kind regards
Arnoud
Please Log in or Create an account to join the conversation.
- siddhartha2020
- Topic Author
- New Member
-

Less
More
- Thank you received: 0
3 years 2 months ago #5
by siddhartha2020
Replied by siddhartha2020 on topic can't perform calibration
I cleaned the machine very well, now it is fine. I used every method. biotinylated protein and aptamers, and his tag proteins. etc... I tried both ways. aptamer immobilization and protein immobilization. I used SA, CM, and NTA chips. we used fluorescence and Au colorimetric assay to check the binding. it was good. but this SPR didn't generate any data. if you know someone who uses this BI4500 biosensing instrument to check aptamers could you please let me know? and you said we can use the ELISA test to check whether immobilization destroys the ligand or not. can you tell how we do that for antibodies( amine coupling)? then I can use that for aptamers. this machine can use for the BSA, their positive control.
Please Log in or Create an account to join the conversation.
- Arnoud
- Moderator
-

Less
More
- Thank you received: 0
3 years 2 months ago - 3 years 2 months ago #6
by Arnoud
Replied by Arnoud on topic can't perform calibration
My first thought was to use the ELISA (coat with protein and come back with aptamer) to confirm that there is actually a binding between protein and aptamer. But since you checked already by other means, this is not necessary any more. What is left is the problem that SPR does not generate any signal between the protein and aptamer. To rule out that a covalent binding of the protein destroys the aptamer binding site, I suggested an ELISA where you covalently bind the protein to the plate. I my mind plates must be available to do that but it turns out that they are not easily available. I found this one:
www.corning.com/catalog/cls/documents/ap...I-MB-101_REV2_DL.pdf
.
Which uses pre-coated plates.
The precoating is the same as you would have after NHS/EDC activation of the carboxyl groups. The example is with amine-modified oligonucleotide (aptamer?) but maybe it is possible to adjust the protocol for a protein. Then you can test if the aptamer still binds to the protein.
Which uses pre-coated plates.
The precoating is the same as you would have after NHS/EDC activation of the carboxyl groups. The example is with amine-modified oligonucleotide (aptamer?) but maybe it is possible to adjust the protocol for a protein. Then you can test if the aptamer still binds to the protein.
Last edit: 3 years 2 months ago by Arnoud.
Please Log in or Create an account to join the conversation.
Moderators: Arnoud, Arnoud